5CCP
 
 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
1JLU
 
 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1PSH
 
 | |
2CYP
 
 | |
2PCB
 
 | |
1FMO
 
 | | CRYSTAL STRUCTURE OF A POLYHISTIDINE-TAGGED RECOMBINANT CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH THE PEPTIDE INHIBITOR PKI(5-24) AND ADENOSINE | | Descriptor: | ADENOSINE, CAMP-DEPENDENT PROTEIN KINASE, HEAT STABLE RABBIT SKELETAL MUSCLE INHIBITOR PROTEIN | | Authors: | Narayana, N, Cox, S, Shaltiel, S, Taylor, S.S, Xuong, N.-H. | | Deposit date: | 1997-07-08 | | Release date: | 1998-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a polyhistidine-tagged recombinant catalytic subunit of cAMP-dependent protein kinase complexed with the peptide inhibitor PKI(5-24) and adenosine.
Biochemistry, 36, 1997
|
|
2PCC
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN ELECTRON TRANSFER PARTNERS, CYTOCHROME C PEROXIDASE AND CYTOCHROME C | | Descriptor: | CYTOCHROME C PEROXIDASE, ISO-1-CYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pelletier, H, Kraut, J. | | Deposit date: | 1993-04-14 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c.
Science, 258, 1992
|
|
1BX6
 
 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1RE8
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 2 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXYBENZOYL)BENZOATE, N-OCTANE, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1REJ
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1REK
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 | | Descriptor: | 3-[(3-SEC-BUTYL-4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXY-5-METHOXYBENZOYL)BENZOATE, PENTANAL, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1HIP
 
 | | TWO-ANGSTROM CRYSTAL STRUCTURE OF OXIDIZED CHROMATIUM HIGH POTENTIAL IRON PROTEIN | | Descriptor: | HIGH POTENTIAL IRON PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Carterjunior, C.W, Kraut, J, Freer, S.T, Xuong, N.-H, Alden, R.A, Bartsch, R.G. | | Deposit date: | 1975-04-01 | | Release date: | 1976-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein.
J.Biol.Chem., 249, 1974
|
|
1RGS
 
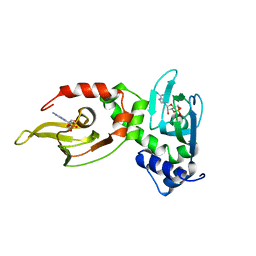 | | REGULATORY SUBUNIT OF CAMP DEPENDENT PROTEIN KINASE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP DEPENDENT PROTEIN KINASE | | Authors: | Su, Y, Dostmann, W.R.G, Herberg, F.W, Durick, K, Xuong, N.-H, Ten Eyck, L, Taylor, S.S, Varughese, K.I. | | Deposit date: | 1995-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains.
Science, 269, 1995
|
|
1BEK
 
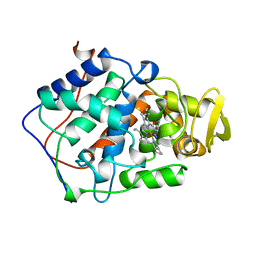 | |
2GLS
 
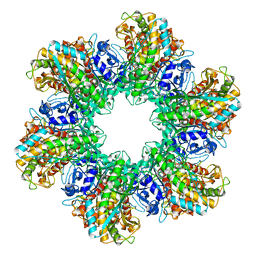 | |
1RL3
 
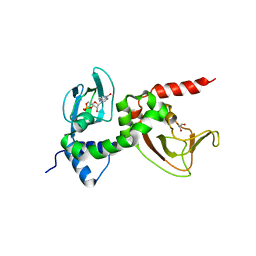 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
2LGS
 
 | | FEEDBACK INHIBITION OF FULLY UNADENYLYLATED GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM BY GLYCINE, ALANINE, AND SERINE | | Descriptor: | GLUTAMIC ACID, GLUTAMINE SYNTHETASE, MANGANESE (II) ION | | Authors: | Liaw, S.-H, Eisenberg, D. | | Deposit date: | 1994-08-05 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1BEP
 
 | | EFFECT OF UNNATURAL HEME SUBSTITUTION ON KINETICS OF ELECTRON TRANSFER IN CYTOCHROME C PEROXIDASE | | Descriptor: | YEAST CYTOCHROME C PEROXIDASE, [7-ETHENYL-12-FORMYL-3,8,13,17-TERTRAMETHYL-21H,23H-PORPHINE-2,18-DIPROPANOATO(2)-N21,N22,N23,N24]IRON | | Authors: | Miller, M, Kraut, J. | | Deposit date: | 1998-05-16 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of Unnatural Heme Substitution on Kinetics of Electron Transfer in Cytochrome C Peroxidase
To be Published
|
|
1VIF
 
 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Narayana, N, Matthews, D.A, Howell, E.E, Xuong, N.-H. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site.
Nat.Struct.Biol., 2, 1995
|
|
1VIE
 
 | |
1BJ9
 
 | | EFFECT OF UNNATURAL HEME SUBSTITUTION ON KINETICS OF ELECTRON TRANSFER IN CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, [7,12-DEACETYL-3,8,13,17-TETRAMETHYL-21H,23H-PORPHINE-2,18-DIPROPANOATO(2-)-N21,N22,N23,N24]-IRON | | Authors: | Miller, M.A, Kraut, J. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of Unnatural Heme Substitution on Kinetics of Electron Transfer in Cytochrome C Peroxidase
To be Published
|
|
1CCP
 
 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
2CEP
 
 | |
1PYG
 
 | |
1NEH
 
 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
