4EUG
 
 | | Crystallographic and Enzymatic Studies of an Active Site Variant H187Q of Escherichia Coli Uracil DNA Glycosylase: Crystal Structures of Mutant H187Q and its Uracil Complex | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187.
Biochemistry, 38, 1999
|
|
6T38
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6T37
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
6TQG
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Glucose-1-phosphate thymidylyltransferase, ... | | Authors: | Alphey, M.S, Xiao, G, Westwood, J.N. | | Deposit date: | 2019-12-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Glucose-1-phosphate thymidylyltransferase (RmlA) inhibitors: An extended SAR study to direct future design.
Bioorg.Med.Chem., 50, 2021
|
|
5EFV
 
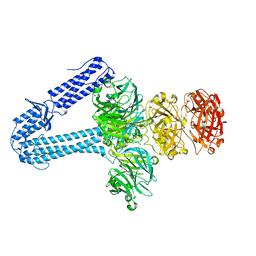 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
4WAC
 
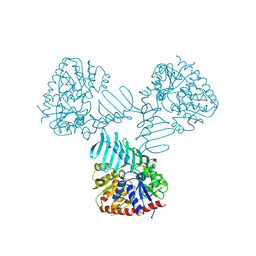 | | Crystal Structure of TarM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koc, C, Stehle, T, Xia, G, Peschel, A. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Enzymatic Analysis of TarM Glycosyltransferase from Staphylococcus aureus Reveals an Oligomeric Protein Specific for the Glycosylation of Wall Teichoic Acid.
J.Biol.Chem., 290, 2015
|
|
4WAD
 
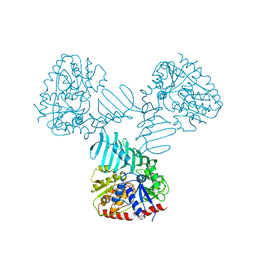 | | Crystal Structure of TarM with UDP-GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glycosyl transferase, ... | | Authors: | Koc, C, Stehle, T, Xia, G, Peschel, A. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Enzymatic Analysis of TarM Glycosyltransferase from Staphylococcus aureus Reveals an Oligomeric Protein Specific for the Glycosylation of Wall Teichoic Acid.
J.Biol.Chem., 290, 2015
|
|
1S2H
 
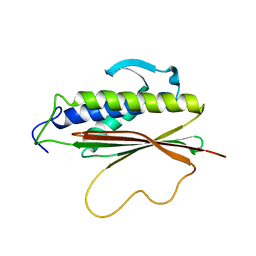 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1NDD
 
 | | STRUCTURE OF NEDD8 | | Descriptor: | CHLORIDE ION, PROTEIN (UBIQUITIN-LIKE PROTEIN NEDD8), SULFATE ION | | Authors: | Whitby, F.G, Xia, G, Pickart, C.M, Hill, C.P. | | Deposit date: | 1998-08-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes.
J.Biol.Chem., 273, 1998
|
|
1FLZ
 
 | | URACIL DNA GLYCOSYLASE WITH UAAP | | Descriptor: | URACIL, URACIL-DNA GLYCOSYLASE | | Authors: | Werner, R.M, Jiang, Y.L, Gordley, R.G, Jagadeesh, G.J, Ladner, J.E, Xiao, G, Tordova, M, Gilliland, G.L, Stivers, J.T. | | Deposit date: | 2000-08-15 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Stressing-out DNA? The contribution of serine-phosphodiester interactions in catalysis by uracil DNA glycosylase.
Biochemistry, 39, 2000
|
|
7A8X
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with the HMA domain of Pikh-1 from rice (Oryza sativa) | | Descriptor: | AVR-Pik protein, NBS-LRR class disease resistance protein | | Authors: | Maidment, J.H.R, Xiao, G, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
1TDJ
 
 | | THREONINE DEAMINASE (BIOSYNTHETIC) FROM E. COLI | | Descriptor: | BIOSYNTHETIC THREONINE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gallagher, D.T, Gilliland, G.L, Xiao, G, Eisenstein, E. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase.
Structure, 6, 1998
|
|
5FWG
 
 | | TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, TETRA-(5-FLUOROTRYPTOPHANYL)-GLUTATHIONE TRANSFERASE MU CLASS | | Authors: | Parsons, J.F, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-11-08 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymes harboring unnatural amino acids: mechanistic and structural analysis of the enhanced catalytic activity of a glutathione transferase containing 5-fluorotryptophan.
Biochemistry, 37, 1998
|
|
1N2A
 
 | | Crystal Structure of a Bacterial Glutathione Transferase from Escherichia coli with Glutathione Sulfonate in the Active Site | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase | | Authors: | Rife, C.L, Parsons, J.F, Xiao, G, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli
Proteins, 53, 2003
|
|
1MTC
 
 | | GLUTATHIONE TRANSFERASE MUTANT Y115F | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, Glutathione S-transferase YB1 | | Authors: | Ladner, J.E, Xiao, G, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Local protein dynamics and catalysis: detection of segmental motion associated with rate-limiting product release by a glutathione transferase
Biochemistry, 41, 2002
|
|
4FDA
 
 | |
1QSG
 
 | | CRYSTAL STRUCTURE OF ENOYL REDUCTASE INHIBITION BY TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, ... | | Authors: | Stewart, M.J, Parikh, S, Xiao, G, Tonge, P.J, Kisker, C. | | Deposit date: | 1999-06-21 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis and mechanism of enoyl reductase inhibition by triclosan.
J.Mol.Biol., 290, 1999
|
|
6H1J
 
 | | TarP native | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable ss-1,3-N-acetylglucosaminyltransferase | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
6H2N
 
 | | TarP-UDP-GlcNAc-Mg | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-13 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
6H21
 
 | | TarP-UDP-GlcNAc-Mn | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
4DZO
 
 | |
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR3
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
2QYF
 
 | |
