3GK2
 
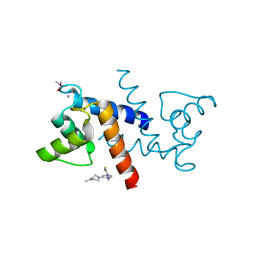 | | X-ray structure of bovine SBi279,Ca(2+)-S100B | | Descriptor: | (Z)-2-[2-(4-methylpiperazin-1-yl)benzyl]diazenecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK4
 
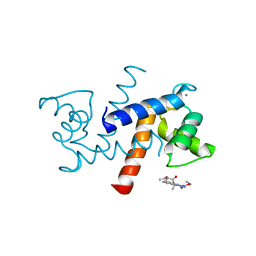 | | X-ray structure of bovine SBi523,Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ethyl 5-{[(1R)-1-(ethoxycarbonyl)-2-oxopropyl]sulfanyl}-1,2-dihydro[1,2,3]triazolo[1,5-a]quinazoline-3-carboxylate | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK1
 
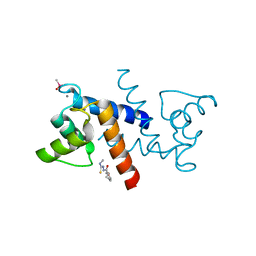 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3IQQ
 
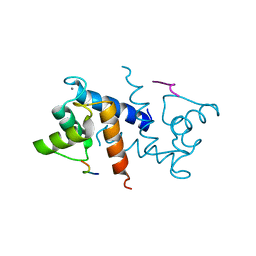 | | X-ray structure of bovine TRTK12-Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, TRTK12 peptide, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-08-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Effects of CapZ Peptide (TRTK-12) Binding to S100B-Ca(2+) as Examined by NMR and X-ray Crystallography
J.Mol.Biol., 396, 2010
|
|
3IQO
 
 | |
3LLE
 
 | | X-ray structure of bovine SC0322,Ca(2+)-S100B | | Descriptor: | 13-methyl-13,14-dihydro[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-28 | | Release date: | 2010-12-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int.J.High Throughput Screen, 2010, 2010
|
|
3LK1
 
 | | X-ray structure of bovine SC0322,Ca(2+)-S100B | | Descriptor: | 2-sulfanylbenzoic acid, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
3LK0
 
 | | X-ray structure of bovine SC0067,Ca(2+)-S100B | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
1C49
 
 | | STRUCTURAL AND FUNCTIONAL DIFFERENCES OF TWO TOXINS FROM THE SCORPION PANDINUS IMPERATOR | | Descriptor: | TOXIN K-BETA | | Authors: | Klenk, K.C, Tenenholz, T.C, Matteson, D.R, Rogowski, R.S, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1999-08-17 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural and functional differences of two toxins from the scorpion Pandinus imperator.
Proteins, 38, 2000
|
|
2K2F
 
 | |
2JUB
 
 | | Solution structure of IPI* | | Descriptor: | Internal protein I | | Authors: | Rifat, D, Wright, N.T, Varney, K.M, Weber, D.J, Black, L.W. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target.
J.Mol.Biol., 375, 2008
|
|
2JVU
 
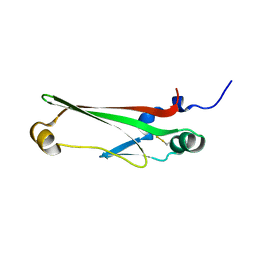 | | Solution Structure of Dispersin from Enteroaggregative Escherichia coli | | Descriptor: | DISPERSIN | | Authors: | Velarde, J.J, Varney, K.M, Farfan, K, Dudley, D, Inman, J.G, Fletcher, J, Weber, D.J, Nataro, J.P. | | Deposit date: | 2007-09-25 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel dispersin protein of enteroaggregative Escherichia coli.
Mol.Microbiol., 66, 2007
|
|
2K7O
 
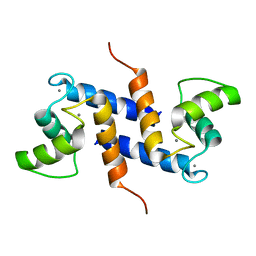 | | Ca2+-S100B, refined with RDCs | | Descriptor: | CALCIUM ION, Protein S100-B | | Authors: | Wright, N.T, Inman, K.G, Levine, J.A, Weber, D.J. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B.
J.Biomol.Nmr, 42, 2008
|
|
1JYT
 
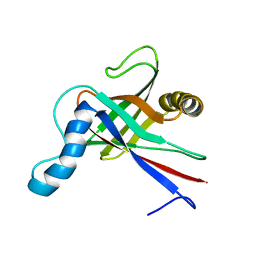 | | Solution structure of olfactory marker protein from rat | | Descriptor: | Olfactory Marker Protein | | Authors: | Baldisseri, D.M, Margolis, J.W, Weber, D.J, Koo, J.H, Margolis, F.M. | | Deposit date: | 2001-09-13 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Olfactory marker protein (OMP) exhibits a beta-clam fold in solution: implications for target peptide interaction and olfactory signal transduction.
J.Mol.Biol., 319, 2002
|
|
1K2H
 
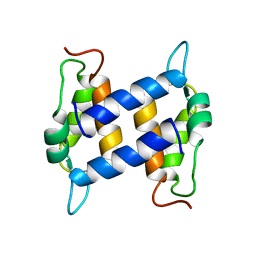 | | Three-dimensional Solution Structure of apo-S100A1. | | Descriptor: | S-100 protein, alpha chain | | Authors: | Rustandi, R.R, Baldisseri, D.M, Inman, K.G, Nizner, P, Hamilton, S.M, Landar, A, Landar, A, Zimmer, D.B, Weber, D.J. | | Deposit date: | 2001-09-27 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR.
Biochemistry, 41, 2002
|
|
2KBM
 
 | | Ca-S100A1 interacting with TRTK12 | | Descriptor: | CALCIUM ION, F-actin-capping protein subunit alpha-2, Protein S100-A1 | | Authors: | Wright, N.T, Varney, K.M, Cannon, B.R, Morgan, M, Weber, D.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ca-S100A1-TRTK12
To be Published
|
|
1M31
 
 | | Three-Dimensional Solution Structure of Apo-Mts1 | | Descriptor: | Placental calcium-binding protein | | Authors: | Vallely, K.M, Rustandi, R.R, Ellis, K.C, Varlamova, O, Bresnick, A.R, Weber, D.J. | | Deposit date: | 2002-06-26 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy.
Biochemistry, 41, 2002
|
|
3RLZ
 
 | |
3RM1
 
 | |
1DT7
 
 | |
8FJ8
 
 | | Crystal structure of Mn(2+),Ca(2+)-S100B | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Protein S100-B | | Authors: | Hunter, D.A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural insights into ions binding to S100A1 versus S100B
To Be Published
|
|
3KO0
 
 | | Structure of the tfp-ca2+-bound activated form of the s100a4 Metastasis factor | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Protein S100-A4 | | Authors: | Malashkevich, V.N, Dulyaninova, N.G, Knight, D, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenothiazines inhibit S100A4 function by inducing protein oligomerization.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8EKL
 
 | |
8EKM
 
 | |
8EKK
 
 | |
