4UDM
 
 | |
6OFR
 
 | |
6U4J
 
 | | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | | Descriptor: | 5-{[(1S)-1-(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]amino}-1-methyl-6-oxo-1,6-dihydropyridine-2-carbonitrile, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-08-25 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6O2Z
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 32 | | Descriptor: | 6-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methylpyridine-3-carbonitrile, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6O2Y
 
 | | Crystal structure of IDH1 R132H mutant in complex with compound 24 | | Descriptor: | 4-{[(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)methyl]amino}-2-methoxybenzonitrile, BETA-MERCAPTOETHANOL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Quinolinone Derivatives as Potent, Selective, and Orally Bioavailable Mutant Isocitrate Dehydrogenase 1 (mIDH1) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6OFS
 
 | |
6OFT
 
 | |
5EAK
 
 | |
8G3W
 
 | | MBP-Mcl1 in complex with ligand 28 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3U
 
 | | MBP-Mcl1 in complex with ligand 21 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-{[(5S,9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]methyl}-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-(ethanediylidene)-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3T
 
 | | MBP-Mcl1 in complex with ligand 12 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3Y
 
 | | MBP-Mcl1 in complex with ligand 34 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3S
 
 | | MBP-Mcl1 in complex with ligand 11 | | Descriptor: | (1'S,3aS,5R,16R,17S,19E,21S,21aR)-6'-chloro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, FORMIC ACID, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3X
 
 | | MBP-Mcl1 in complex with ligand 32 | | Descriptor: | 1,2-ETHANEDIOL, Maltodextrin-binding protein, Induced myeloid leukemia cell differentiation protein Mcl-1 chimera, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
3IFO
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:10D5) complex | | Descriptor: | 10D5 FAB antibody heavy chain, 10D5 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3IFN
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-40:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3IFP
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12B4) complex | | Descriptor: | 12B4 FAB antibody heavy chain, 12B4 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3IFL
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
4IWD
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | Descriptor: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
5ODW
 
 | | Structure of the FpvAI-pyocin S2 complex | | Descriptor: | Ferripyoverdine receptor, Pyocin-S2, SULFATE ION | | Authors: | White, P, Joshi, A, Kleanthous, C. | | Deposit date: | 2017-07-06 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exploitation of an iron transporter for bacterial protein antibiotic import.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5EW5
 
 | |
5VTG
 
 | |
4I1H
 
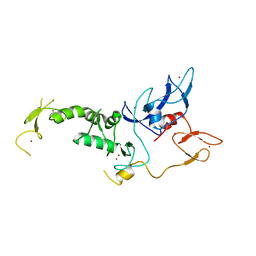 | | Structure of Parkin E3 ligase | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
4I1F
 
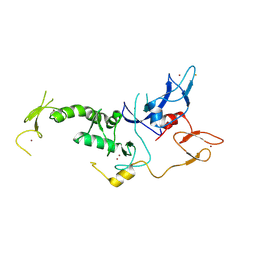 | | Structure of Parkin-S223P E3 ligase | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases.
Nat Commun, 4, 2013
|
|
7PVC
 
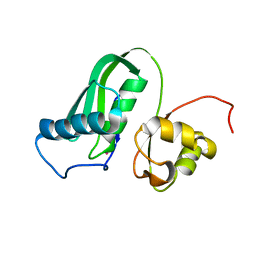 | | The structure of Kbp.K from E. coli with potassium bound. | | Descriptor: | POTASSIUM ION, Potassium binding protein Kbp | | Authors: | Smith, B.O. | | Deposit date: | 2021-10-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-11 | | Method: | SOLUTION NMR | | Cite: | Tuning the Sensitivity of Genetically Encoded Fluorescent Potassium Indicators through Structure-Guided and Genome Mining Strategies.
ACS Sens, 7, 2022
|
|
