3UET
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
1QB4
 
 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | Descriptor: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | Deposit date: | 1999-04-30 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
6W0V
 
 | | The Crystal Structure of the Mutant Nuclease Domain of Pyocin S8 with its Cognate Immunity Protein | | Descriptor: | Bacteriocin immunity protein, Pyocin S8 | | Authors: | Turano, H, Gomes, F, Domingos, R.M, Netto, L.E.S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Molecular Structure and Functional Analysis of Pyocin S8 from Pseudomonas aeruginosa Reveals the Essential Requirement of a Glutamate Residue in the H-N-H Motif for DNase Activity.
J.Bacteriol., 202, 2020
|
|
3ICP
 
 | |
5Z6B
 
 | | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, Putative ABC transporter substrate-binding protein YesO | | Authors: | Sugiura, H, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide
To Be Published
|
|
5Z6C
 
 | |
3KO8
 
 | | Crystal Structure of UDP-galactose 4-epimerase | | Descriptor: | NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Sakuraba, H, Kawai, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UDP-galactose 4-epimerase from the hyperthermophilic archaeon Pyrobaculum calidifontis
Arch.Biochem.Biophys., 512, 2011
|
|
3STL
 
 | | KcsA potassium channel mutant Y82C with Cadmium bound | | Descriptor: | CADMIUM ION, POTASSIUM ION, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
4JJG
 
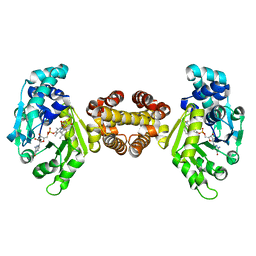 | | Crystal structure of FE-hydrogenase from methanothermobacter marburgensis in complex with toluenesulfonylmethylisocyanide | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, N-methyl-1-[(4-methylbenzyl)sulfonyl]methanamine, iron-guanylyl pyridinol cofactor | | Authors: | Tamura, H, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3STZ
 
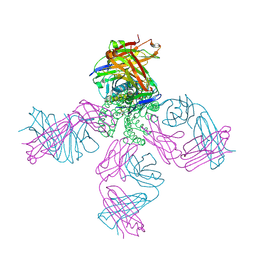 | | KcsA potassium channel mutant Y82C with nitroxide spin label | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
4JJF
 
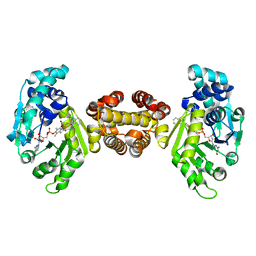 | | Crystal structure of FE-hydrogenase from methanothermobacter marburgensis in complex with 2-naphthylisocyanide | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, N-(naphthalen-2-yl)methanimine, iron-guanylyl pyridinol cofactor | | Authors: | Tamura, H, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3PHF
 
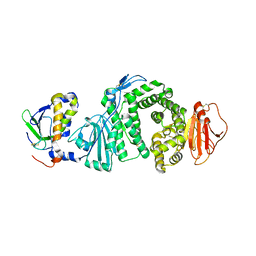 | |
1MHO
 
 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
3E9I
 
 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysine hydroxamate-AMP | | Descriptor: | 5'-O-{(R)-hydroxy[(L-lysylamino)oxy]phosphoryl}adenosine, Lysyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
3E9H
 
 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysylsulfamoyl adenosine | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
1K1X
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1K1W
 
 | | Crystal structure of 4-alpha-glucanotransferase from thermococcus litoralis | | Descriptor: | 4-ALPHA-GLUCANOTRANSFERASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1K1Y
 
 | | Crystal structure of thermococcus litoralis 4-alpha-glucanotransferase complexed with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-ALPHA-GLUCANOTRANSFERASE, ... | | Authors: | Imamura, H, Fushinobu, S, Kumasaka, T, Yamamoto, M, Jeon, B.S, Wakagi, T, Matsuzawa, H. | | Deposit date: | 2001-09-26 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor
J.BIOL.CHEM., 278, 2003
|
|
1VCV
 
 | |
2YVK
 
 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis | | Descriptor: | 5-S-METHYL-1-O-PHOSPHONO-5-THIO-D-RIBULOSE, Methylthioribose-1-phosphate isomerase | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2YRF
 
 | | Crystal structure of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis complexed with sulfate ion | | Descriptor: | Methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Tamura, H, Inoue, T, Kai, Y, Matsumura, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: Implications for catalytic mechanism
Protein Sci., 17, 2008
|
|
2CW7
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2ZVI
 
 | | Crystal structure of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis | | Descriptor: | 2,3-diketo-5-methylthiopentyl-1-phosphate enolase | | Authors: | Tamura, H, Yadani, T, Kai, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2008-11-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3VTF
 
 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
