3HRS
 
 | | Crystal Structure of the Manganese-activated Repressor ScaR: apo form | | Descriptor: | Metalloregulator ScaR, SULFATE ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3HRU
 
 | | Crystal Structure of ScaR with bound Zn2+ | | Descriptor: | Metalloregulator ScaR, SULFATE ION, ZINC ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
4OFQ
 
 | | Structure of the C-terminal domain of the Streptococcus pyogenes antigen I/II-family protein AspA | | Descriptor: | CALCIUM ION, Putative cell surface protein | | Authors: | Hall, M, Nylander, A, Jenkinson, H.F, Persson, K. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of AspA (antigen I/II-family) protein from Streptococcus pyogenes.
FEBS Open Bio, 4, 2014
|
|
5ZRD
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at low pH condition | | Descriptor: | CITRIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5ZRE
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at high pH condition | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN ATOM, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
6K8W
 
 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6K8S
 
 | |
6K8U
 
 | |
6K8V
 
 | |
6K8T
 
 | |
6IJ6
 
 | |
6IJ3
 
 | |
6IJ5
 
 | |
6IJ4
 
 | |
5X7M
 
 | |
5X5T
 
 | |
5X7N
 
 | |
5X5U
 
 | |
6ABY
 
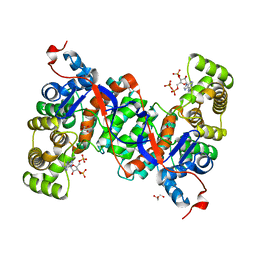 | | Crystal structure of citrate synthase (Msed_1522) from Metallosphaera sedula in complex with oxaloacetate | | Descriptor: | ACETYL COENZYME *A, Citrate synthase, GLYCEROL, ... | | Authors: | Lee, S.-H, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the inhibition properties of archaeon citrate synthase from Metallosphaera sedula.
PLoS ONE, 14, 2019
|
|
5ITS
 
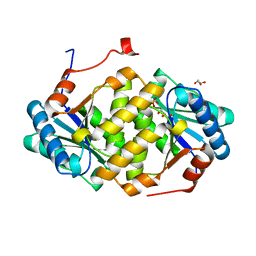 | | Crystal structure of LOG from Corynebacterium glutamicum | | Descriptor: | Cytokinin riboside 5'-monophosphate phosphoribohydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Seo, H.-G, Kim, K.-J. | | Deposit date: | 2016-03-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for cytokinin production by LOG from Corynebacterium glutamicum
Sci Rep, 6, 2016
|
|
2C5L
 
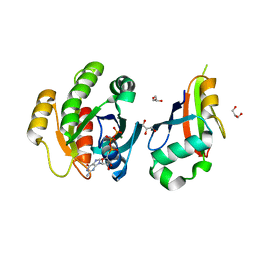 | | Structure of PLC epsilon Ras association domain with hRas | | Descriptor: | GLYCEROL, GTPASE HRAS, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roe, S.M, Bunney, T.D, Katan, M, Pearl, L.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon
Mol.Cell, 21, 2006
|
|
6ABX
 
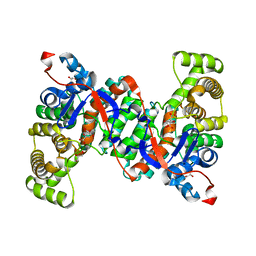 | |
4NZS
 
 | |
7DZV
 
 | |
7DZT
 
 | | Cyrstal structure of PETase from Rhizobacter gummiphilus | | Descriptor: | DLH domain-containing protein | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Implications for the PET decomposition mechanism through similarity and dissimilarity between PETases from Rhizobacter gummiphilus and Ideonella sakaiensis.
J Hazard Mater, 416, 2021
|
|
