1R6X
 
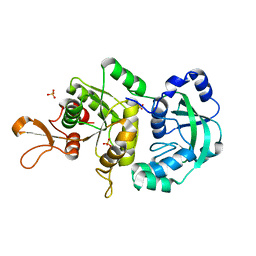 | | The Crystal Structure of a Truncated Form of Yeast ATP Sulfurylase, Lacking the C-Terminal APS Kinase-like Domain, in complex with Sulfate | | Descriptor: | ATP:sulfate adenylyltransferase, COBALT (II) ION, SULFATE ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity
Protein Eng., 16, 2003
|
|
1T57
 
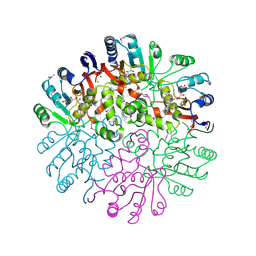 | | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved Protein MTH1675, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION | | Authors: | Kim, Y, Joachimiak, A, Saridakis, V, Xu, X, Arrowsmith, C.H, Christendat, D, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Conserved Protein MTH1675 from Methanobacterium thermoautotrophicum
To be Published
|
|
1Q1L
 
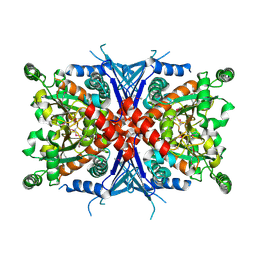 | | Crystal Structure of Chorismate Synthase | | Descriptor: | Chorismate synthase | | Authors: | Viola, C.M, Saridakis, V, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-07-21 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of chorismate synthase from Aquifex aeolicus reveals a novel beta alpha beta sandwich topology
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1EP0
 
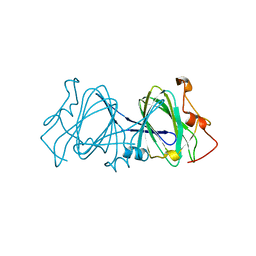 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EPZ
 
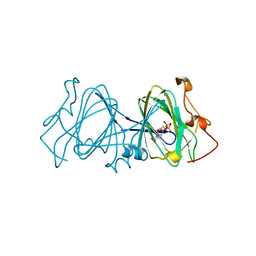 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1L1S
 
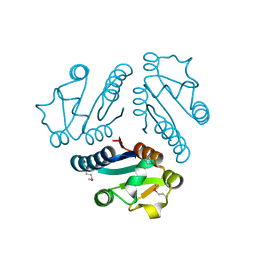 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
4YP7
 
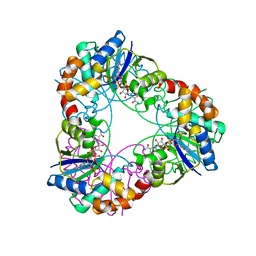 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YP6
 
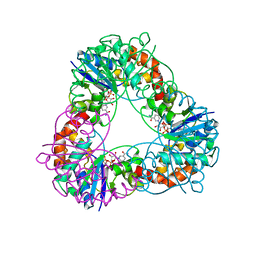 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YP5
 
 | | Crystal structure of Methanobacterium thermoautotrophicum NMNAT in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Pfoh, R, Christendat, D, Pai, E.F, Saridakis, V. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Nicotinamide mononucleotide adenylyltransferase displays alternate binding modes for nicotinamide nucleotides.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4YSI
 
 | | Structure of USP7 with a novel viral protein | | Descriptor: | GLYCEROL, SER-PRO-GLY-GLU-GLY-PRO-SER-GLY, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Chavoshi, S, Saridakis, V. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of USP7 with a novel viral protein
J.Biol.Chem., 2016
|
|
1GH9
 
 | |
1EIJ
 
 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
2OB9
 
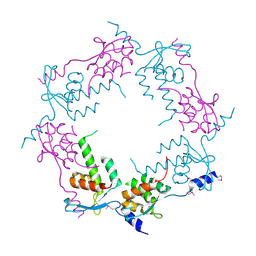 | | Structure of bacteriophage HK97 tail assembly chaperone | | Descriptor: | Tail assembly chaperone | | Authors: | McGrath, T.E, Tuite, A, Bona, D, Saridakis, V, Edwards, A.M, Maxwell, K, Chirgadze, N.Y. | | Deposit date: | 2006-12-18 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved spiral structure for highly diverged phage tail assembly chaperones.
J.Mol.Biol., 425, 2013
|
|
4IS7
 
 | |
2FH0
 
 | |
