4D2B
 
 | | Structure of a ligand free POT family peptide transporter | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, DI-OR TRIPEPTIDE:H+ SYMPORTER, ... | | Authors: | Lyons, J.A, Parker, J.L, Solcan, N, Brinth, A, Li, D, Shah, S.T.A, Caffrey, M, Newstead, S. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Polyspecificity in the Pot Family of Proton-Coupled Oligopeptide Transporters.
Embo Rep., 15, 2014
|
|
4D2C
 
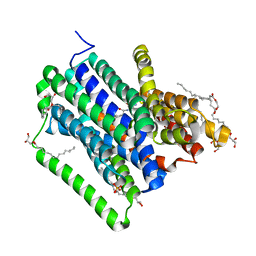 | | Structure of a di peptide bound POT family peptide transporter | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ALANINE, ... | | Authors: | Lyons, J.A, Parker, J.L, Solcan, N, Brinth, A, Li, D, Shah, S.T.A, Caffrey, M, Newstead, S. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Basis for Polyspecificity in the Pot Family of Proton-Coupled Oligopeptide Transporters.
Embo Rep., 15, 2014
|
|
4D2D
 
 | | Structure of a tri peptide bound POT family peptide transporter | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ALANINE-TRIPEPTIDE, ... | | Authors: | Lyons, J.A, Parker, J.L, Solcan, N, Brinth, A, Li, D, Shah, S.T.A, Caffrey, M, Newstead, S. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural Basis for Polyspecificity in the Pot Family of Proton-Coupled Oligopeptide Transporters.
Embo Rep., 15, 2014
|
|
1OB9
 
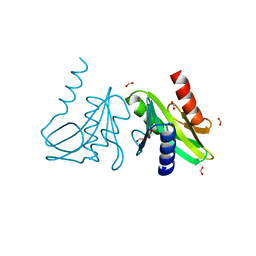 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, HOLLIDAY JUNCTION RESOLVASE | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
1OB8
 
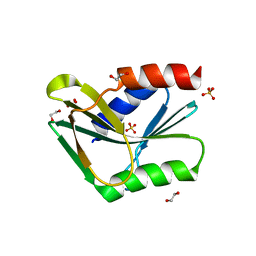 | | Holliday Junction Resolving Enzyme | | Descriptor: | 1,2-ETHANEDIOL, HOLLIDAY-JUNCTION RESOLVASE, SULFATE ION | | Authors: | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | Deposit date: | 2003-01-28 | | Release date: | 2004-10-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
6EI3
 
 | | Crystal structure of auto inhibited POT family peptide transporter | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Proton-dependent oligopeptide transporter family protein | | Authors: | Newstead, S, Brinth, A, Vogeley, L, Caffrey, M. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proton movement and coupling in the POT family of peptide transporters.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7OYE
 
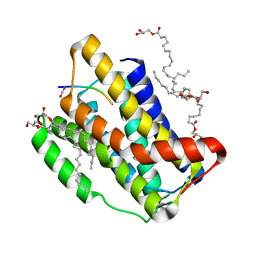 | |
6F34
 
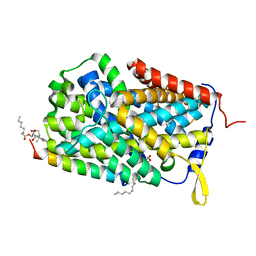 | |
5OQT
 
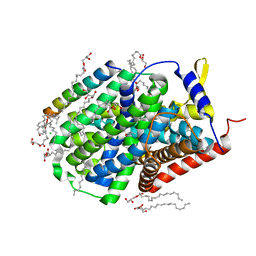 | |
4TKK
 
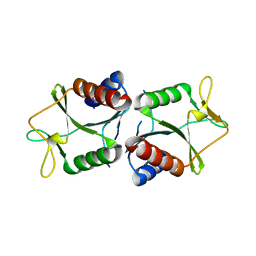 | | Sulfolobus solfataricus HJC mutants | | Descriptor: | Holliday junction resolvase Hjc | | Authors: | Bond, C.S. | | Deposit date: | 2014-05-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Unengineering: Reducing the Crystallisability of Sulfolobus solfataricus Hjc
Aust.J.Chem., 67, 2014
|
|
4TKD
 
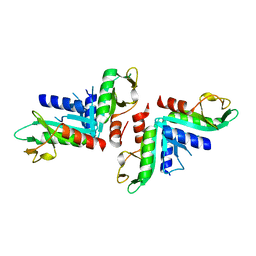 | | Sulfolobus solfataricus HJC mutants | | Descriptor: | Holliday junction resolvase Hjc | | Authors: | Bond, C.S. | | Deposit date: | 2014-05-26 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Unengineering: Reducing the Crystallisability of Sulfolobus solfataricus Hjc
Aust.J.Chem., 67, 2014
|
|
6I6J
 
 | |
6I6B
 
 | |
6I6H
 
 | |
6Y7V
 
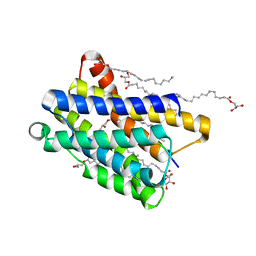 | | Crystal structure of the KDEL receptor bound to HDEL peptide at pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON DIOXIDE, ER lumen protein-retaining receptor 2, ... | | Authors: | Braeuer, P, Newstead, S. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | A signal capture and proofreading mechanism for the KDEL-receptor explains selectivity and dynamic range in ER retrieval.
Elife, 10, 2021
|
|
7ZKZ
 
 | |
7ZK1
 
 | |
5A9I
 
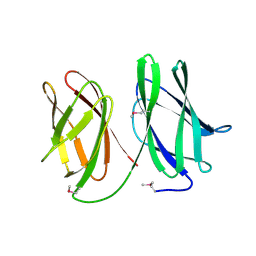 | | Crystal structure of the extracellular domain of PepT2 | | Descriptor: | CITRIC ACID, GLYCEROL, SOLUTE CARRIER FAMILY 15 MEMBER 2 | | Authors: | Beale, J.H, Newstead, S. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
5A9H
 
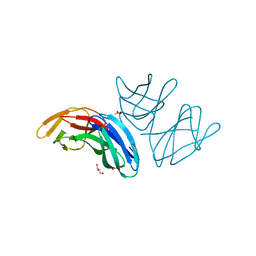 | | Crystal structure of the extracellular domain of PepT2 | | Descriptor: | ACETATE ION, CESIUM ION, GLYCEROL, ... | | Authors: | Beale, J.H, Newstead, S. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
5A9D
 
 | | Crystal structure of the extracellular domain of PepT1 | | Descriptor: | GLYCEROL, SOLUTE CARRIER FAMILY 15 MEMBER 1 | | Authors: | Beale, J.H, Bird, L.E, Owens, R.J, Newstead, S. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
3VS2
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Parker, J.L, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
