3PCK
 
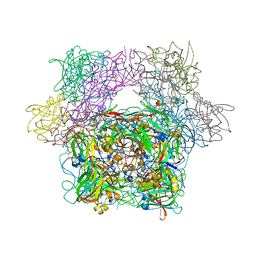 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 6-HYDROXYNICOTINIC ACID N-OXIDE | | Descriptor: | 6-HYDROXYISONICOTINIC ACID N-OXIDE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding.
Biochemistry, 36, 1997
|
|
3PCE
 
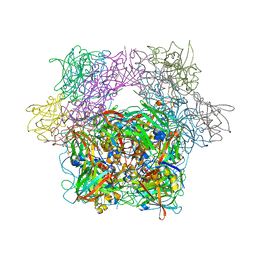 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-HYDROXYPHENYLACETATE | | Descriptor: | 3-HYDROXYPHENYLACETATE, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Elango, N, Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-04-29 | | Release date: | 1998-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3EA6
 
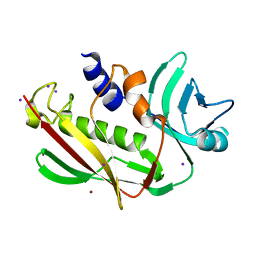 | | Atomic resolution of crystal structure of SEK | | Descriptor: | IODIDE ION, Staphylococcal enterotoxin K, ZINC ION | | Authors: | Shi, K, Huseby, M, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2008-08-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Studies of an Emerging Pyrogenic Superantigen, SEK
To be Published
|
|
5TSS
 
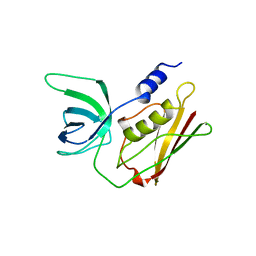 | | TOXIC SHOCK SYNDROME TOXIN-1: ORTHORHOMBIC P222(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
1TS2
 
 | | T128A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
3I41
 
 | |
3I48
 
 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound magnesium ions | | Descriptor: | Beta-hemolysin, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
to be published, 2009
|
|
3I5V
 
 | |
3I46
 
 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound calcium ions | | Descriptor: | Beta-hemolysin, CALCIUM ION, CHLORIDE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
To be Published
|
|
4P2P
 
 | | AN INDEPENDENT CRYSTALLOGRAPHIC REFINEMENT OF PORCINE PHOSPHOLIPASE A2 AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Finzel, B.C, Ohlendorf, D.H, Weber, P.C, Salemme, F.R. | | Deposit date: | 1991-10-22 | | Release date: | 1992-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An independent crystallographic refinement of porcine phospholipase A2 at 2.4 A resolution
Acta Crystallogr.,Sect.B, 47, 1991
|
|
2GRM
 
 | | Crystal structure of PrgX/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
2GRL
 
 | | Crystal structure of dCT/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
4BP2
 
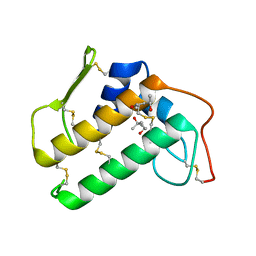 | | CRYSTALLOGRAPHIC REFINEMENT OF BOVINE PRO-PHOSPHOLIPASE A2 AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Finzel, B.C, Weber, P.C, Ohlendorf, D.H, Salemme, F.R. | | Deposit date: | 1990-09-07 | | Release date: | 1991-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic refinement of bovine pro-phospholipase A2 at 1.6 A resolution.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1XF1
 
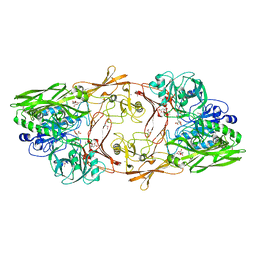 | | Structure of C5a peptidase- a key virulence factor from Streptococcus | | Descriptor: | ACETATE ION, C5a peptidase, CALCIUM ION, ... | | Authors: | Brown, C.K, Gu, Z.Y, Cleary, P.P, Matsuka, Y, Olmstead, S, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2004-09-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the streptococcal cell wall C5a peptidase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YKP
 
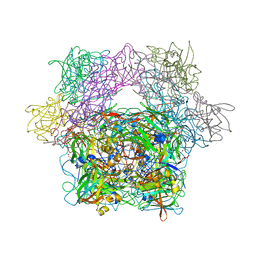 | | Protocatechuate 3,4-Dioxygenase Y408H mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
3HL6
 
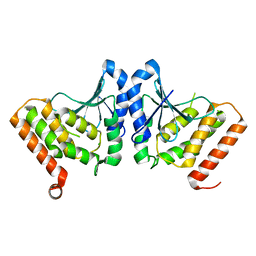 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
3JS1
 
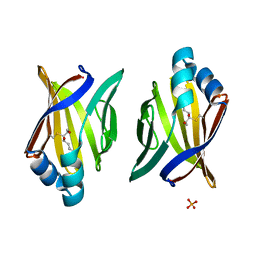 | | Crystal structure of adipocyte fatty acid binding protein covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | Adipocyte fatty acid-binding protein, PHOSPHATE ION | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
1MHY
 
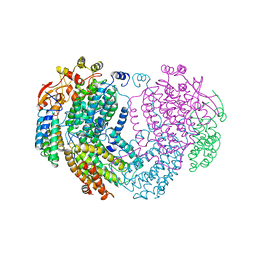 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
1MHZ
 
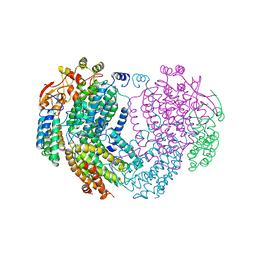 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
1EO2
 
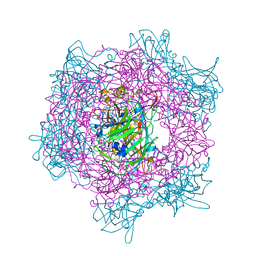 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, PROTOCATECHUATE 3,4-DIOXYGENASE BETA CHAIN | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-21 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EO9
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE AT PH < 7.0 | | Descriptor: | FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, PROTOCATECHUATE 3,4-DIOXYGENASE BETA CHAIN;, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EOB
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EOC
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH 4-NITROCATECHOL | | Descriptor: | 4-NITROCATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
1EOA
 
 | | CRYSTAL STRUCTURE OF ACINETOBACTER SP. ADP1 PROTOCATECHUATE 3,4-DIOXYGENASE IN COMPLEX WITH CYANIDE | | Descriptor: | CYANIDE ION, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, D'Argenio, D.A, Ornston, L.N, Ohlendorf, D.H. | | Deposit date: | 2000-03-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 A resolution: implications for the mechanism of an intradiol dioxygenase.
Biochemistry, 39, 2000
|
|
