2CPI
 
 | | Solution structure of the RNA recognition motif of CNOT4 | | Descriptor: | CCR4-NOT transcription complex subunit 4 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif of CNOT4
To be Published
|
|
2CPE
 
 | | Solution structure of the RNA recognition motif of Ewing Sarcoma(EWS) protein | | Descriptor: | RNA-binding protein EWS | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA recognition motif of Ewing Sarcoma(EWS) protein
To be Published
|
|
2CPH
 
 | | Solution structure of the C-terminal RNA recognition motif of hypothetical RNA-binding protein RBM19 | | Descriptor: | RNA binding motif protein 19 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal RNA recognition motif of hypothetical RNA-binding protein RBM19
To be Published
|
|
2CPM
 
 | | Solution structure of the R3H domain of human sperm-associated antigen 7 | | Descriptor: | Sperm-associated antigen 7 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the R3H domain of human sperm-associated antigen 7
To be Published
|
|
2GGF
 
 | | Solution structure of the MA3 domain of human Programmed cell death 4 | | Descriptor: | Programmed cell death 4, isoform 1 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-24 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MA3 domain of human Programmed cell death 4
To be Published
|
|
2CPF
 
 | | Solution structure of the penultimate RNA recognition motif of hypothetical RNA-binding protein RBM19 | | Descriptor: | RNA binding motif protein 19 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the penultimate RNA recognition motif of hypothetical RNA-binding protein RBM19
To be Published
|
|
2CPR
 
 | | Solution structure of the HRDC domain of human Exosome component 10 | | Descriptor: | Exosome component 10 | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Exosome component 10
To be Published
|
|
2CPQ
 
 | | Solution structure of the N-terminal KH domain of human FXR1 | | Descriptor: | Fragile X mental retardation syndrome related protein 1, isoform b' | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal KH domain of human FXR1
To be Published
|
|
2EBI
 
 | | Arabidopsis GT-1 DNA-binding domain with T133D phosphomimetic mutation | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Noto, K, Niyada, E, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
5H7U
 
 | | NMR structure of eIF3 36-163 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit C | | Authors: | Nagata, T, Obayashi, E. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
7BV9
 
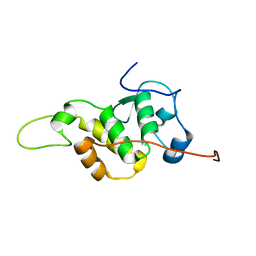 | | The NMR structure of the BEN domain from human NAC1 | | Descriptor: | Nucleus accumbens-associated protein 1 | | Authors: | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
2RVH
 
 | | NMR structure of eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Nagata, T, Obayashi, E, Asano, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
6J3G
 
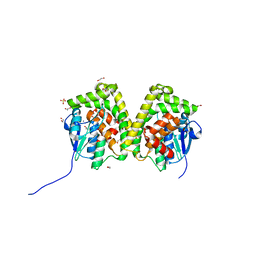 | | Crystal structure of an apo form of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutathione S-transferase, ... | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
6J3H
 
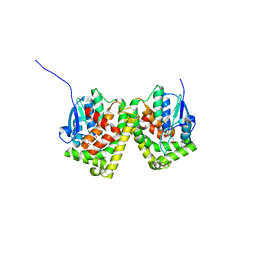 | | Crystal structure of the glutathione S-transferase, CsGST83044, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of key residues for activities of atypical glutathione S-transferase of Ceriporiopsis subvermispora, a selective degrader of lignin in woody biomass, by crystallography and functional mutagenesis.
Int.J.Biol.Macromol., 132, 2019
|
|
6J3F
 
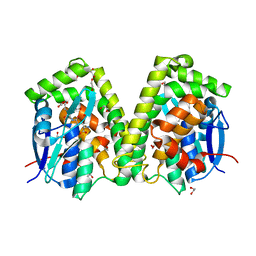 | | Crystal structure of the glutathione S-transferase, CsGST63524, of Ceriporiopsis subvermispora in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, glutathione S-transferase | | Authors: | Osman, W.H.W, Mikami, B, Saka, N, Kondo, K, Nagata, T, Katahira, M. | | Deposit date: | 2019-01-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a serine-type glutathione S-transferase of Ceriporiopsis subvermispora and identification of the enzymatically important non-canonical residues by functional mutagenesis.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
8WLS
 
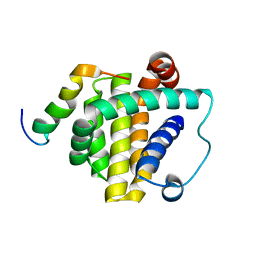 | |
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
7EXK
 
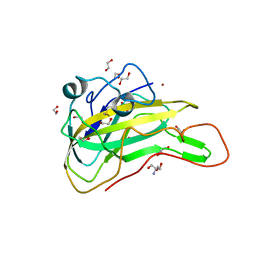 | | An AA9 LPMO of Ceriporiopsis subvermispora | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nguyen, H, Kondo, K, Nagata, T, Katahira, M, Mikami, B. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional and Structural Characterizations of Lytic Polysaccharide Monooxygenase, Which Cooperates Synergistically with Cellulases, from Ceriporiopsis subvermispora.
Acs Sustain Chem Eng, 10, 2022
|
|
8JH8
 
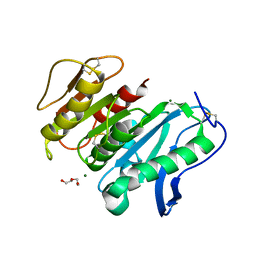 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Feruloyl esterase, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
8JH9
 
 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Feruloyl esterase with ferulic acid, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
1UAW
 
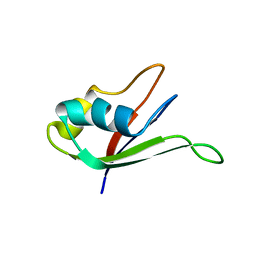 | | Solution structure of the N-terminal RNA-binding domain of mouse Musashi1 | | Descriptor: | mouse-musashi-1 | | Authors: | Miyanoiri, Y, Kobayashi, H, Watanabe, M, Ikeda, T, Nagata, T, Okano, H, Uesugi, S, Katahira, M. | | Deposit date: | 2003-03-24 | | Release date: | 2004-03-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Origin of higher affinity to RNA of the N-terminal RNA-binding domain than that of the C-terminal one of a mouse neural protein, musashi1, as revealed by comparison of their structures, modes of interaction, surface electrostatic potentials, and backbone dynamics
J.Biol.Chem., 278, 2003
|
|
8IYB
 
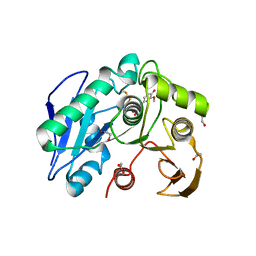 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IYC
 
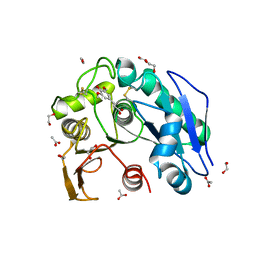 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IY8
 
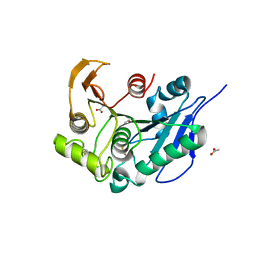 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Feruloyl esterase | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
