2VRK
 
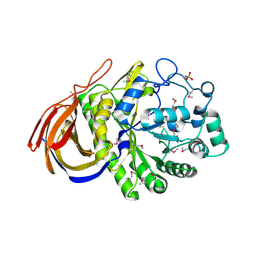 | | Structure of a seleno-methionyl derivative of wild type arabinofuranosidase from Thermobacillus xylanilyticus | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE, PHOSPHATE ION | | Authors: | Paes, G, Skov, L.K, ODonohue, M.J, Remond, C, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Complex between a Branched Pentasaccharide and Thermobacillus Xylanilyticus Gh-51 Arabinofuranosidase Reveals Xylan-Binding Determinants and Induced Fit.
Biochemistry, 47, 2008
|
|
2VRQ
 
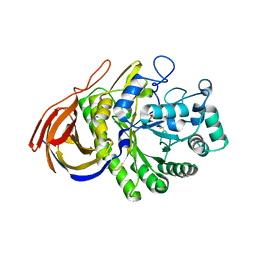 | | STRUCTURE OF AN INACTIVE MUTANT OF ARABINOFURANOSIDASE FROM THERMOBACILLUS XYLANILYTICUS IN COMPLEX WITH A PENTASACCHARIDE | | Descriptor: | ALPHA-L-ARABINOFURANOSIDASE, PHOSPHATE ION, alpha-L-arabinofuranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Paes, G, Skov, L.K, Odonohue, M.J, Remond, C, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Complex between a Branched Pentasaccharide and Thermobacillus Xylanilyticus Gh-51 Arabinofuranosidase Reveals Xylan-Binding Determinants and Induced Fit.
Biochemistry, 47, 2008
|
|
1LLT
 
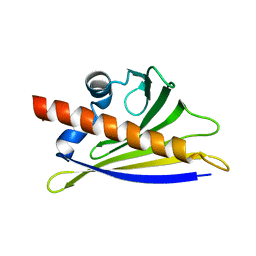 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT E45S | | Descriptor: | POLLEN ALLERGEN BET V 1 | | Authors: | Spangfort, M.D, Mirza, O, Ipsen, H, Van Neerven, R.J, Gajhede, M, Larsen, J.N. | | Deposit date: | 2002-04-30 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis.
J.Immunol., 171, 2003
|
|
2CMO
 
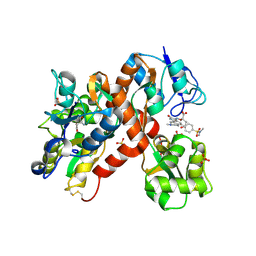 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | Descriptor: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
2B76
 
 | | E. coli Quinol fumarate reductase FrdA E49Q mutation | | Descriptor: | CITRATE ANION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Maklashina, E, Iverson, T.M, Sher, Y, Kotlyar, V, Mirza, O, Andrell, J, Hudson, J.M, Armstrong, F.A, Cecchini, G. | | Deposit date: | 2005-10-03 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fumarate Reductase and Succinate Oxidase Activity of Escherichia coli Complex II Homologs Are Perturbed Differently by Mutation of the Flavin Binding Domain
J.Biol.Chem., 281, 2006
|
|
1R7A
 
 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
4AYS
 
 | | The Structure of Amylosucrase from D. radiodurans | | Descriptor: | AMYLOSUCRASE | | Authors: | Skov, L.K, Pizzut, S, Remaud-Simeon, M, Gajhede, M, Mirza, O. | | Deposit date: | 2012-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Structure of Amylosucrase from Deinococcus Radiodurans Has an Unusual Open Active-Site Topology
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6RUQ
 
 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
5M99
 
 | | Functional Characterization and Crystal Structure of Thermostable Amylase from Thermotoga petrophila, reveals High Thermostability and an Archaic form of Dimerization | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-amylase, ... | | Authors: | Hameed, U, Price, I, Mirza, O.A. | | Deposit date: | 2016-11-01 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Biochim. Biophys. Acta, 1865, 2017
|
|
1PA2
 
 | | ARABIDOPSIS THALIANA PEROXIDASE A2 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PEROXIDASE, ... | | Authors: | Henriksen, A. | | Deposit date: | 1999-05-18 | | Release date: | 2000-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification.
Plant Mol.Biol., 44, 2000
|
|
1GWO
 
 | |
1GX2
 
 | |
1GWT
 
 | |
1QNX
 
 | | Ves v 5, an allergen from Vespula vulgaris venom | | Descriptor: | SODIUM ION, VES V 5 | | Authors: | Henriksen, A, Gajhede, M, Spangfort, M.D. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-26 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Major Venom Allergen of Yellow Jackets, Ves V 5: Structural Characterization of a Pathogenesis-Related Protein Superfamily.
Proteins: Struct.,Funct., Genet., 45, 2001
|
|
