7AU1
 
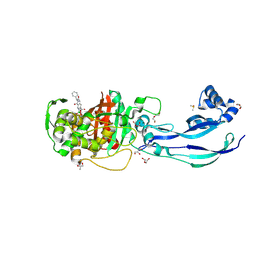 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 12) | | Descriptor: | 2-(6-(((R)-2-amino-2-oxo-1-phenylethyl)carbamoyl)-1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acetic acid, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
1XDV
 
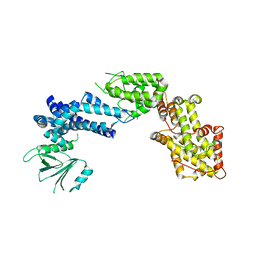 | | Experimentally Phased Structure of Human the Son of Sevenless protein at 4.1 Ang. | | Descriptor: | Son of sevenless protein homolog 1 | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XD4
 
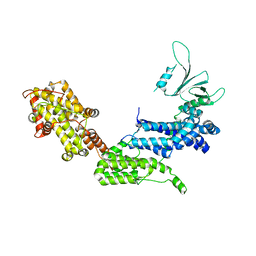 | | Crystal structure of the DH-PH-cat module of Son of Sevenless (SOS) | | Descriptor: | Son of sevenless protein homolog 1 | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XD2
 
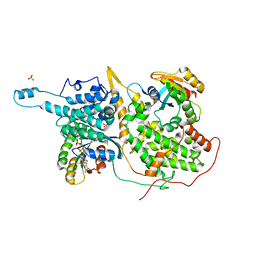 | | Crystal Structure of a ternary Ras:SOS:Ras*GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
5I69
 
 | | MBP-MamC magnetite-interaction component mutant-D70A | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Tercedor, C.V, Kolusheva, S, Gonzalez, T.P, Widdrat, M, Grimberg, N, Levi, H, Nelkenbaum, O, Davidove, G, Faivre, D, Jimenez-Lopez, C, Zarivach, R. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC.
J.Struct.Biol., 194, 2016
|
|
2FGT
 
 | | Crystal Structure of YycH from Bacillus subtilis | | Descriptor: | Two-component system yycF/yycG regulatory protein yycH | | Authors: | Szurmant, H, Zhao, H, Mohan, M.A, James, H.A, Varughese, K.I. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of YycH involved in the regulation of the essential YycFG two-component system in Bacillus subtilis reveals a novel tertiary structure.
Protein Sci., 15, 2006
|
|
6QRU
 
 | | X-ray radiation dose series on xylose isomerase - 2.01 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRY
 
 | | X-ray radiation dose series on xylose isomerase - merged data | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRT
 
 | | X-ray radiation dose series on xylose isomerase - 1.38 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRR
 
 | | X-ray radiation dose series on xylose isomerase - 0.13 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.096 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRV
 
 | | X-ray radiation dose series on xylose isomerase - 2.63 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRX
 
 | | X-ray radiation dose series on xylose isomerase - 3.88 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRS
 
 | | X-ray radiation dose series on xylose isomerase - 0.13 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
6QRW
 
 | | X-ray radiation dose series on xylose isomerase - 3.25 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
3PXK
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH Pyrrolo[2,3-d]thiazole | | Descriptor: | 6-(4,4-dimethylpent-2-ynyl)-4~{H}-pyrrolo[2,3-d][1,3]thiazole, PTK2 protein, SULFATE ION | | Authors: | Koolman, H, Heinrich, T, Musil, D. | | Deposit date: | 2010-12-10 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Co-crystal Structures of FAK with an Unprecedented Pyrrolo[2,3-d]thiazole
To be Published
|
|
1SSO
 
 | | SOLUTION STRUCTURE AND DNA-BINDING PROPERTIES OF A THERMOSTABLE PROTEIN FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | SSO7D | | Authors: | Baumann, H, Knapp, S, Lundback, T, Ladenstein, R, Hard, T. | | Deposit date: | 1995-03-31 | | Release date: | 1995-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus.
Nat.Struct.Biol., 1, 1994
|
|
3STZ
 
 | | KcsA potassium channel mutant Y82C with nitroxide spin label | | Descriptor: | POTASSIUM ION, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
5A02
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol | | Descriptor: | ALDOSE-ALDOSE OXIDOREDUCTASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
4L79
 
 | | Crystal Structure of nucleotide-free Myosin 1b residues 1-728 with bound Calmodulin | | Descriptor: | Calmodulin, MAGNESIUM ION, Unconventional myosin-Ib | | Authors: | Shuman, H, Zwolak, A, Dominguez, R, Ostap, E.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MH0
 
 | | Selective activation of Epac1 and Epac2 | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-{6-amino-8-[(4-fluorobenzyl)sulfanyl]-9H-purin-9-yl}-2-sulfanyltetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-7-ol 2-oxide, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
4JX8
 
 | |
4MGK
 
 | | Selective activation of Epac1 and Epac2 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
4MGZ
 
 | | Selective activation of Epac1 and Epac2 | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-[6-amino-8-(benzylsulfanyl)-9H-purin-9-yl]-2-sulfanyltetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-7-ol 2-oxide, Rap guanine nucleotide exchange factor 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective activation of Epac1 and Epac2
To be Published
|
|
4JQC
 
 | | Crystal Structure of E.coli Enoyl Reductase in Complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI, N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Subramanya, H, Rao, K.N, Anirudha, L. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E.coli Enoyl Reductase in Complex with NAD and AFN-1252
TO BE PUBLISHED
|
|
5A04
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
