8Z78
 
 | |
8Z6P
 
 | |
8JCS
 
 | |
8OH9
 
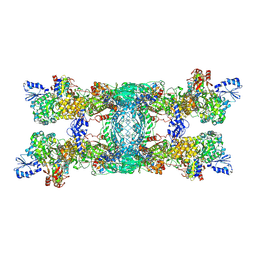 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
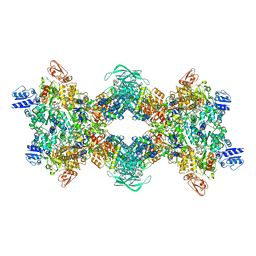 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
6JMI
 
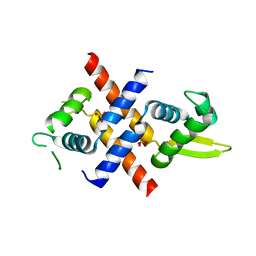 | | Crystal structure of M.tuberculosis Rv0081 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv0081 | | Authors: | Kumar, A, Phulera, S, Mande, C.S. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-10 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of hypoxic gene regulation by the Rv0081 transcription factor of Mycobacterium tuberculosis.
Febs Lett., 593, 2019
|
|
7CXD
 
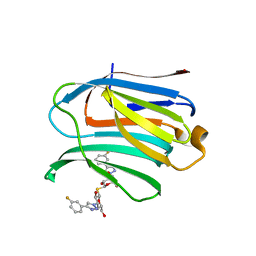 | | Xray structure of rat Galectin-3 CRD in complex with TD-139 belonging to P121 space group | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, BROMIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXC
 
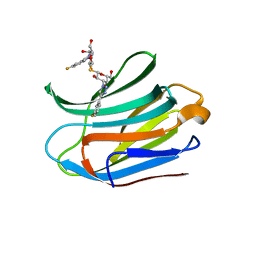 | |
7CXA
 
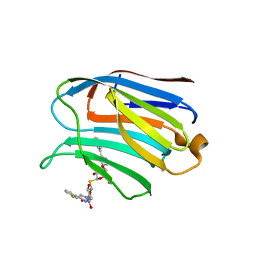 | | Structure of human Galectin-3 CRD in complex with TD-139 belonging to P31 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXB
 
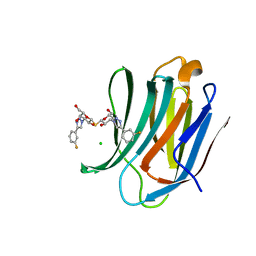 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7RBK
 
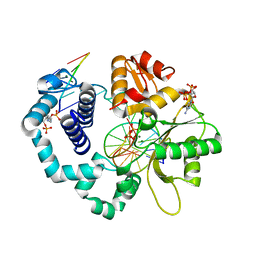 | | Human DNA polymerase beta crosslinked complex, 40 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7M6M
 
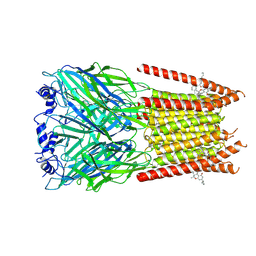 | | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6N
 
 | |
7M6Q
 
 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6R
 
 | |
7M6S
 
 | |
7M6O
 
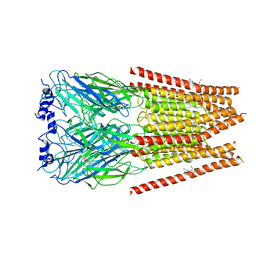 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6P
 
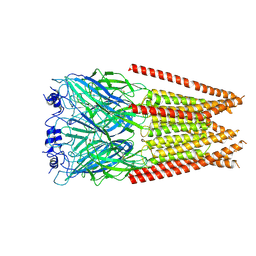 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7RBJ
 
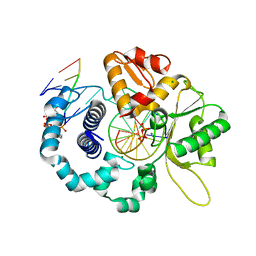 | | Human DNA polymerase beta crosslinked complex, 30 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBN
 
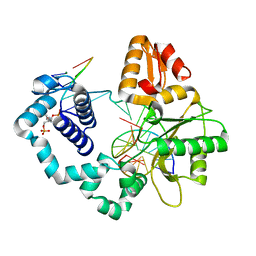 | | Human DNA polymerase beta crosslinked complex, 20 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBI
 
 | | Human DNA polymerase beta crosslinked complex, 20 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBO
 
 | | Human DNA polymerase beta crosslinked complex, 60 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBM
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mn exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBH
 
 | | Human DNA polymerase beta crosslinked ternary complex 2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBL
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
