1JOV
 
 | | Crystal Structure Analysis of HI1317 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HI1317, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Howard, A.J, Eisenstein, E, Gilliland, G, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-31 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal 1.57-A Crystal Structure of HI1317
TO BE PUBLISHED
|
|
1I3H
 
 | | CONCANAVALIN A-DIMANNOSE STRUCTURE | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Sanders, D.A.R, Moothoo, D.N, Raftery, J, Howard, A.J, Helliwell, J.R, Naismith, J.H. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution structure of the Con A-dimannose complex.
J.Mol.Biol., 310, 2001
|
|
1I2L
 
 | | DEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI WITH INHIBITOR | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1I2K
 
 | | AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jensen, P.Y, Parsons, J.F, Fisher, K.E, Pachikara, A.S, Tordova, M, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-02-09 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Mechanism of Escherichia coli Aminodeoxychorismate Lyase
To be Published
|
|
1JJV
 
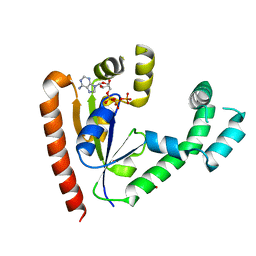 | | DEPHOSPHO-COA KINASE IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEPHOSPHO-COA KINASE, MERCURY (II) ION, ... | | Authors: | Obmolova, G, Teplyakov, A, Bonander, N, Eisenstein, E, Howard, A.J, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae.
J.Struct.Biol., 136, 2001
|
|
1LQA
 
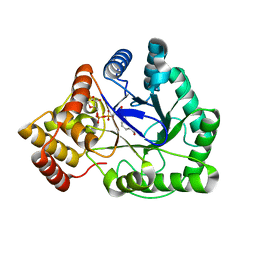 | | TAS PROTEIN FROM ESCHERICHIA COLI IN COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tas protein | | Authors: | Obmolova, G, Teplyakov, A, Khil, P.P, Howard, A.J, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1S01
 
 | |
1AK9
 
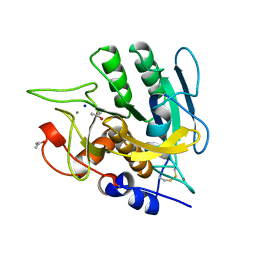 | | SUBTILISIN MUTANT 8321 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-05-30 | | Release date: | 1997-11-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1A2Q
 
 | | SUBTILISIN BPN' MUTANT 7186 | | Descriptor: | ACETONE, CALCIUM ION, SUBTILISIN BPN' | | Authors: | Gilliland, G.L, Whitlow, M, Howard, A.J. | | Deposit date: | 1998-01-08 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AU9
 
 | | SUBTILISIN BPN' MUTANT 8324 IN CITRATE | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN BPN', ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-09-12 | | Release date: | 1997-12-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AQN
 
 | | SUBTILISIN MUTANT 8324 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN 8324, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-07-31 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stabilityfor subtilisin BPN' through incremental changes in the free energy of unfolding
To be published
|
|
3CA8
 
 | | Crystal structure of Escherichia coli YdcF, an S-adenosyl-L-methionine utilizing enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein ydcF, SULFATE ION | | Authors: | Lim, K, Chao, K, Lehmann, C, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes.
Proteins, 72, 2008
|
|
1M65
 
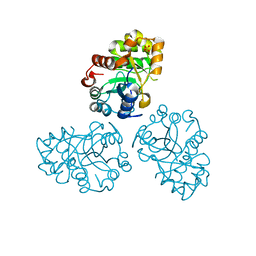 | | YCDX PROTEIN | | Descriptor: | Hypothetical protein ycdX, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-07-12 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M68
 
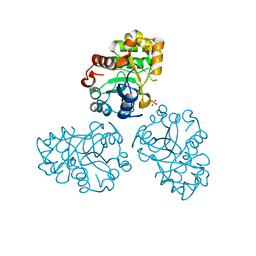 | | YCDX PROTEIN, TRINUCLEAR ZINC SITE | | Descriptor: | Hypothetical protein ycdX, SULFATE ION, ZINC ION | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L. | | Deposit date: | 2002-07-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a
trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
3CPP
 
 | |
2CPP
 
 | |
1PHD
 
 | |
1PHC
 
 | |
1PHG
 
 | |
1PHF
 
 | |
1PHE
 
 | |
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-10-21 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
2O0O
 
 | | Crystal structure of TL1A | | Descriptor: | MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-11-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
1PHB
 
 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
1PHA
 
 | | INHIBITOR-INDUCED CONFORMATIONAL CHANGE IN CYTOCHROME P450-CAM | | Descriptor: | 1-(N-IMIDAZOLYL)-2-HYDROXY-2-(2,3-DICHLOROPHENYL)OCTANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Poulos, T.L. | | Deposit date: | 1992-07-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Inhibitor-induced conformational change in cytochrome P-450CAM.
Biochemistry, 32, 1993
|
|
