2D9Q
 
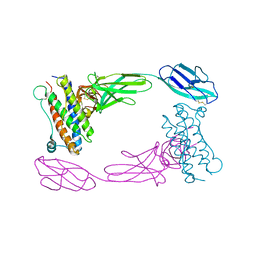 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5XSW
 
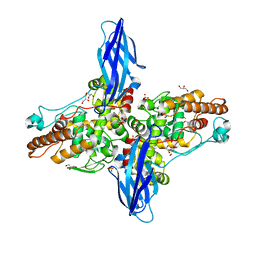 | |
5XSV
 
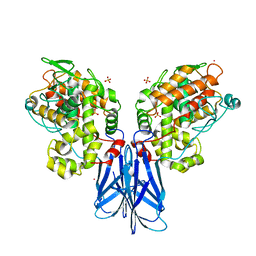 | |
5XSX
 
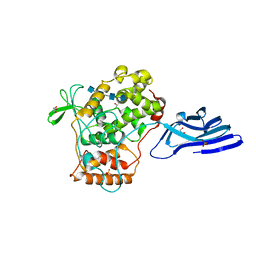 | | Crystal structure of an archaeal chitinase in the substrate-complex form (P212121) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2017-06-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of an archaeal chitinase ChiD and its ligand complexes.
Glycobiology, 28, 2018
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1AUV
 
 | |
1AUX
 
 | |
1IJT
 
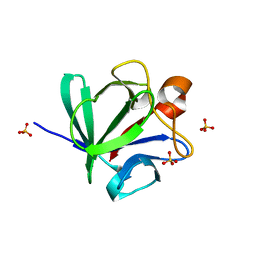 | | Crystal Structure of Fibroblast Growth Factor 4 (FGF4) | | Descriptor: | SULFATE ION, fibroblast growth factor 4 | | Authors: | Bellosta, P, Plotnikov, A.N, Eliseenkova, A.V, Basilico, C, Mohammadi, M. | | Deposit date: | 2001-04-29 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of receptor and heparin binding sites in fibroblast growth factor 4 by structure-based mutagenesis.
Mol.Cell.Biol., 21, 2001
|
|
2EK8
 
 | |
2EK9
 
 | |
