3TJ1
 
 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | Descriptor: | RNA polymerase I-specific transcription initiation factor RRN3 | | Authors: | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
6YN1
 
 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
5HYM
 
 | | 3-Hydroxybenzoate 6-hydroxylase from Rhodococcus jostii in complex with phosphatidylinositol | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Phosphatidylinositol, ... | | Authors: | Orru, R, Montersino, S, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Hydroxybenzoate 6-Hydroxylase from Rhodococcus jostii RHA1 Contains a Phosphatidylinositol Cofactor.
Front Microbiol, 8, 2017
|
|
3T8X
 
 | | Crystal structure of human CD1b in complex with synthetic antigenic diacylsulfoglycolipid SGL12 and endogenous spacer | | Descriptor: | 2-O-sulfo-alpha-D-glucopyranosyl 2-O-hexadecanoyl-3-O-[(2E,4S,6S,8S)-2,4,6,8-tetramethyltetracos-2-enoyl]-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Mourey, L, Julien, S. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the human CD1b Antigen-binding groove for presentation of mycobacterial sulfoglycolipids
To be Published
|
|
7OK5
 
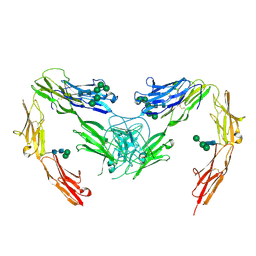 | | Crystal structure of mouse neurofascin 155 immunoglobulin domains | | Descriptor: | Neurofascin 155, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
7OL4
 
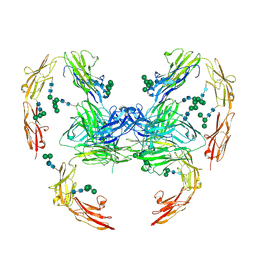 | |
5LB7
 
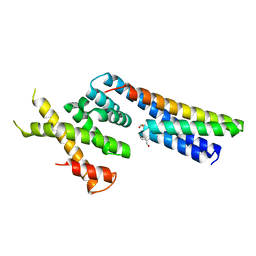 | | Complex structure between p60N/p80C katanin and a peptide derived from ASPM | | Descriptor: | Abnormal spindle-like microcephaly-associated protein homolog, Katanin p60 ATPase-containing subunit A1, Katanin p80 WD40 repeat-containing subunit B1 | | Authors: | Rezabkova, L, Capitani, G, Kammerer, R.A, Steinmetz, M.O. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Nat. Cell Biol., 19, 2017
|
|
5NMT
 
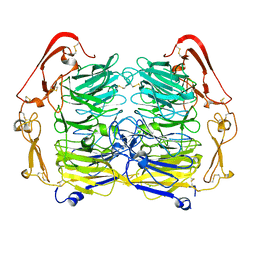 | | Dimer structure of Sortilin ectodomain crystal form 1, 2.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
6ZLY
 
 | |
5N8Y
 
 | | KaiCBA circadian clock backbone model based on a Cryo-EM density | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Schuller, J.M, Snijder, J, Loessl, P, Heck, A.J.R, Foerster, F. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state.
Science, 355, 2017
|
|
5NNJ
 
 | | Dimer structure of Sortilin ectodomain crystal form 3, 4.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2017-04-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
5NNI
 
 | | Dimer structure of Sortilin ectodomain crystal form 2, 3.2 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2017-04-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
