8SDN
 
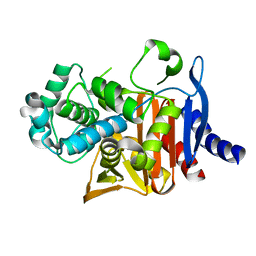 | | Crystal structure of PDC-3 Y221H beta-lactamase | | Descriptor: | Beta-lactamase, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8SDR
 
 | |
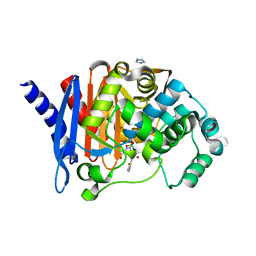
8SDT
 
 | |
8SDL
 
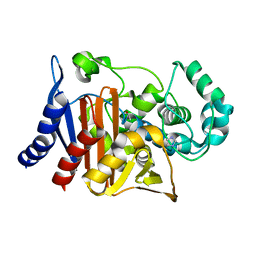 | | Crystal structure of PDC-3 beta-lactamase | | Descriptor: | Beta-lactamase, IMIDAZOLE, ISOPROPYL ALCOHOL | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Natural protein engineering in the Omega-loop: the role of Y221 in ceftazidime and ceftolozane resistance in Pseudomonas -derived cephalosporinase.
Antimicrob.Agents Chemother., 67, 2023
|
|
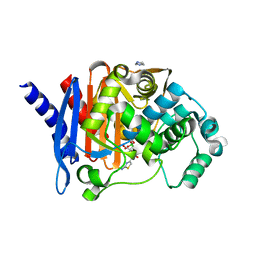
8SDS
 
 | |
6HKY
 
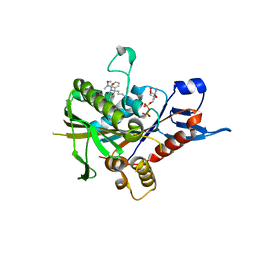 | | Eg5-inhibitor complex | | Descriptor: | (2~{S})-2-(3-azanylpropyl)-5-[2,5-bis(fluoranyl)phenyl]-~{N}-methoxy-~{N}-methyl-2-phenyl-1,3,4-thiadiazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-09-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Is the Fate of Clinical Candidate Arry-520 Already Sealed? Predicting Resistance in Eg5-Inhibitor Complexes.
Mol.Cancer Ther., 18, 2019
|
|
6HKX
 
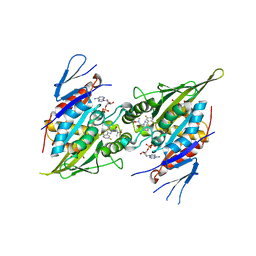 | | Eg5-inhibitor complex | | Descriptor: | (2~{S})-2-(3-azanylpropyl)-5-[2,5-bis(fluoranyl)phenyl]-~{N}-methoxy-~{N}-methyl-2-phenyl-1,3,4-thiadiazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-09-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is the Fate of Clinical Candidate Arry-520 Already Sealed? Predicting Resistance in Eg5-Inhibitor Complexes.
Mol.Cancer Ther., 18, 2019
|
|
8CRF
 
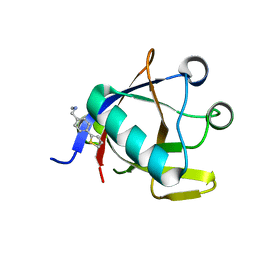 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | Descriptor: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | Authors: | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRK
 
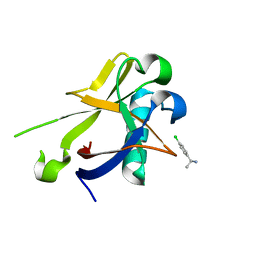 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRM
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11C6 refined against anomalous diffraction data | | Descriptor: | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
