1MJ0
 
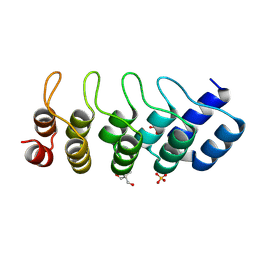 | | SANK E3_5: an artificial Ankyrin repeat protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SANK E3_5 Protein, SULFATE ION | | Authors: | Kohl, A, Binz, H.K, Forrer, P, Stumpp, M.T, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-08-26 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Designed to be stable: Crystal structure of a consensus ankyrin repeat protein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SBN
 
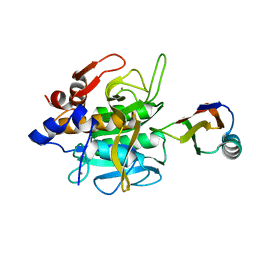 | |
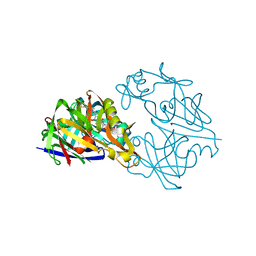
1RNE
 
 | |
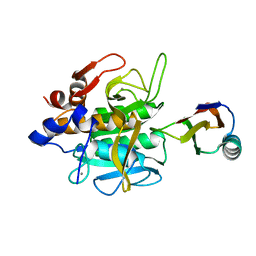
1SIB
 
 | |
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
1N3N
 
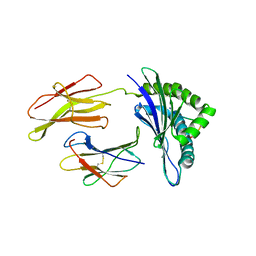 | | Crystal structure of a mycobacterial hsp60 epitope with the murine class I MHC molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Capitani, G, Tissot, A.C, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2002-10-29 | | Release date: | 2003-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of mycobacterial
and murine hsp60 epitopes in
complex with the class I MHC
molecule H-2D(b)
FEBS Lett., 543, 2003
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4BPQ
 
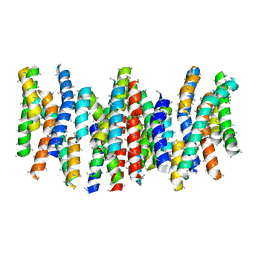 | | Structure and substrate induced conformational changes of the secondary citrate-sodium symporter CitS revealed by electron crystallography | | Descriptor: | CITRATE:SODIUM SYMPORTER | | Authors: | Kebbel, F, Kurz, M, Arheit, M, Gruetter, M.G, Stahlberg, H. | | Deposit date: | 2013-05-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Structure and Substrate-Induced Conformational Changes of the Secondary Citrate/Sodium Symporter Cits Revealed by Electron Crystallography.
Structure, 21, 2013
|
|
4LSZ
 
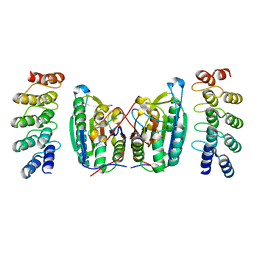 | | Caspase-7 in Complex with DARPin D7.18 | | Descriptor: | Caspase-7 subunit p10, Caspase-7 subunit p20, DARPin D7.18 | | Authors: | Fluetsch, A, Lukarska, M, Gruetter, M.G. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combined inhibition of caspase 3 and caspase 7 by two highly selective DARPins slows down cellular demise.
Biochem.J., 461, 2014
|
|
3LZM
 
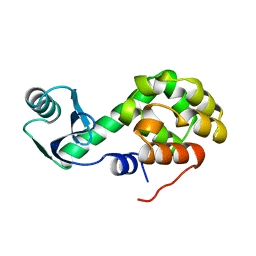 | |
1F9E
 
 | | CASPASE-8 SPECIFICITY PROBED AT SUBSITE S4: CRYSTAL STRUCTURE OF THE CASPASE-8-Z-DEVD-CHO | | Descriptor: | (PHQ)DEVD, CASPASE-8 ALPHA CHAIN, CASPASE-8 BETA CHAIN | | Authors: | Blanchard, H, Donepudi, M, Tschopp, M, Kodandapani, L, Wu, J.C, Grutter, M.G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-07-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-cho complex.
J.Mol.Biol., 302, 2000
|
|
2J8S
 
 | | Drug Export Pathway of Multidrug Exporter AcrB Revealed by DARPin Inhibitors | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, DARPIN, DODECYL-ALPHA-D-MALTOSIDE, ... | | Authors: | Sennhauser, G, Amstutz, P, Briand, C, Storchenegger, O, Gruetter, M.G. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Drug Export Pathway of Multidrug Exporter Acrb Revealed by Darpin Inhibitors.
Plos Biol., 5, 2007
|
|
2K7Z
 
 | |
2MIB
 
 | |
2LZM
 
 | |
4HJ0
 
 | | Crystal structure of the human GIPr ECD in complex with Gipg013 Fab at 3-A resolution | | Descriptor: | Gastric inhibitory polypeptide receptor, Gipg013 Fab, Antagonizing antibody to the GIP Receptor, ... | | Authors: | Madhurantakam, C, Ravn, P, Gruetter, M.G, Jackson, R.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Pharmacological Characterization of Novel Potent and Selective Monoclonal Antibody Antagonists of Glucose-dependent Insulinotropic Polypeptide Receptor.
J.Biol.Chem., 288, 2013
|
|
4Q4A
 
 | | Improved model of AMP-PNP bound TM287/288 | | Descriptor: | ABC transporter, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q4J
 
 | | Structure of crosslinked TM287/288_S498C_S520C mutant | | Descriptor: | ABC transporter, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Hohl, M, Schoeppe, J, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q7K
 
 | |
4Q7L
 
 | | Structure of NBD288 of TM287/288 | | Descriptor: | NICKEL (II) ION, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Bukowska, M.A, Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Transporter Motor Taken Apart: Flexibility in the Nucleotide Binding Domains of a Heterodimeric ABC Exporter.
Biochemistry, 54, 2015
|
|
4Q4H
 
 | | TM287/288 in its apo state | | Descriptor: | ABC transporter, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q7M
 
 | | Structure of NBD288-Avi of TM287/288 | | Descriptor: | Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Bukowska, M.A, Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Transporter Motor Taken Apart: Flexibility in the Nucleotide Binding Domains of a Heterodimeric ABC Exporter.
Biochemistry, 54, 2015
|
|
1PII
 
 | |
