5L7V
 
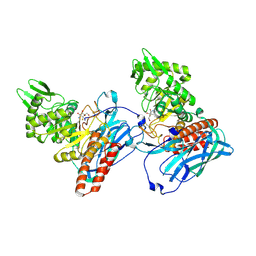 | | Crystal Structure of BvGH123 with bond transition state analog Galthiazoline. | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
5L7U
 
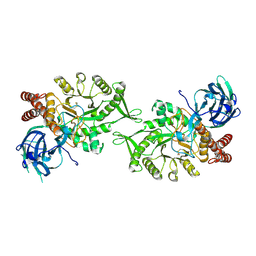 | | Crystal structure of BvGH123 with bound GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, Glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
5L7R
 
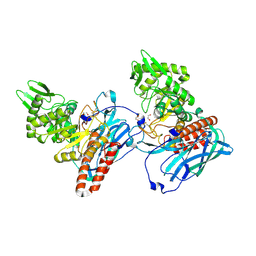 | | Crystal structure of BvGH123 | | Descriptor: | 1,2-ETHANEDIOL, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
5JTS
 
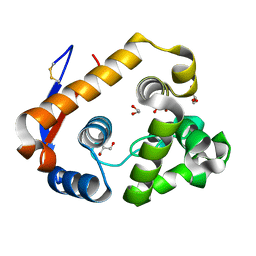 | | Structure of a beta-1,4-mannanase, SsGH134. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
3VPL
 
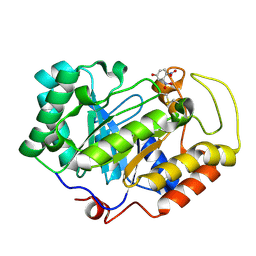 | | Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase | | Descriptor: | 3,4-dinitrophenol, Beta-1,3-xylanase XYL4, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-2-deoxy-2-fluoro-beta-D-xylopyranose, ... | | Authors: | Watanabe, N, Sakaguchi, K. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 beta-1,3-xylanase at 1.2 A resolution
To be Published
|
|
5TIH
 
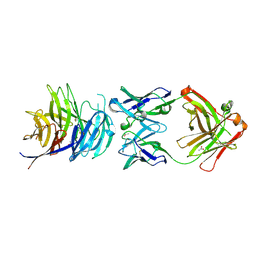 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | ACETATE ION, CyRPA antibody Fab Heavy Chain, CyRPA antibody Fab Light Chain, ... | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5TIK
 
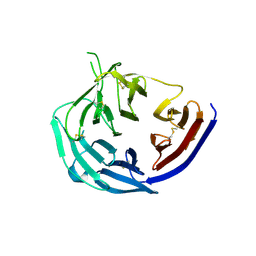 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | Cysteine-rich protective antigen | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5OHT
 
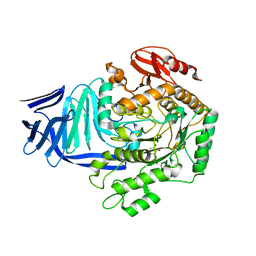 | | A GH31 family sulfoquinovosidase from E. coli in complex with aza-sugar inhibitor IFGSQ | | Descriptor: | CALCIUM ION, Sulfoquinovosidase, [(3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)piperidin-3-yl]methanesulfonic acid | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
5OHS
 
 | | A GH31 family sulfoquinovosidase mutant D455N in complex with pNPSQ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-nitrophenyl alpha-D-6-sulfoquinovoside, ... | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
5OHY
 
 | | A GH31 family sulfoquinovosidase in complex with aza-sugar inhibitor IFGSQ | | Descriptor: | 1,2-ETHANEDIOL, Alpha-glucosidase yihQ, PHOSPHATE ION, ... | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
6VLF
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
8S5B
 
 | | Crystal structure of the sulfoquinovosyl binding protein (SmoF) from A. tumefaciens sulfo-SMO pathway in complex with SQOctyl ligand | | Descriptor: | Sulfoquinovosyl glycerol-binding protein SmoF, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-octoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Sharma, M, Davies, G.J. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture-and-release of a sulfoquinovose-binding protein on sulfoquinovose-modified agarose.
Org.Biomol.Chem., 22, 2024
|
|
6SMY
 
 | |
6SMZ
 
 | |
6SM7
 
 | | Crystal structure of SLA Reductase YihU from E. Coli | | Descriptor: | 3-sulfolactaldehyde reductase, BORIC ACID | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dynamic Structural Changes Accompany the Production of Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase
Acs Catalysis, 2020
|
|
7QHV
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with sulfoquinovosyl diacylglycerol | | Descriptor: | GLYCINE, Sulfoquinovosyl binding protein, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-[(2~{S})-3-butanoyloxy-2-heptanoyloxy-propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A, Davies, G.J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7OLF
 
 | |
7YZS
 
 | |
7YZU
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with SQMe | | Descriptor: | Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-methoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Davies, G.J. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7NE2
 
 | | Crystal structure of class I SFP aldolase YihT from Salmonella enterica with SFP/ DHAP (Schiff base complex with active site Lys193) | | Descriptor: | (2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)-6-phosphonooxy-hexane-1-sulfonic acid, Sulfofructosephosphate aldolase, [(~{E})-2,3-bis(oxidanyl)prop-1-enyl] dihydrogen phosphate | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
7NBZ
 
 | |
7OH2
 
 | |
7OFX
 
 | | Crystal structure of a GH31 family sulfoquinovosidase mutant D455N from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | Alpha-glucosidase yihQ, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8Q59
 
 | | Crystal structure of metal-dependent class II sulfofructose phosphate aldolase from Yersinia aldovae in complex with sulfofructose phosphate (YaSqiA-Zn-SFP) | | Descriptor: | (3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-oxidanylidene-6-phosphonooxy-hexane-1-sulfonic acid, SODIUM ION, Tagatose-1,6-bisphosphate aldolase kbaY, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
