1PMT
 
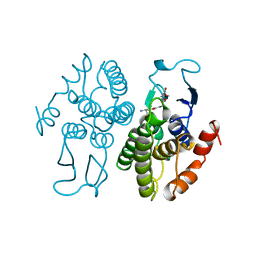 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-03-23 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
1MWP
 
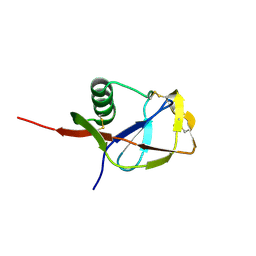 | | N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN | | Descriptor: | AMYLOID A4 PROTEIN | | Authors: | Rossjohn, J, Cappai, R, Feil, S.C, Henry, A, McKinstry, W.J, Galatis, D, Hesse, L, Multhaup, G, Beyreuther, K, Masters, C.L, Parker, M.W. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein.
Nat.Struct.Biol., 6, 1999
|
|
1FHE
 
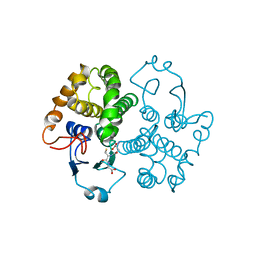 | | GLUTATHIONE TRANSFERASE (FH47) FROM FASCIOLA HEPATICA | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization, structural determination and analysis of a novel parasite vaccine candidate: Fasciola hepatica glutathione S-transferase.
J.Mol.Biol., 273, 1997
|
|
3SL5
 
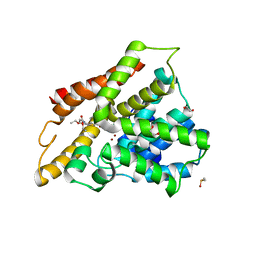 | | Crystal structure of the catalytic domain of PDE4D2 complexed with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL4
 
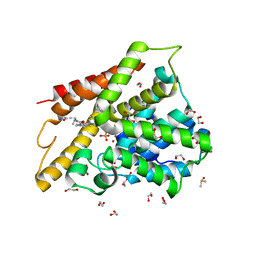 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10D | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL6
 
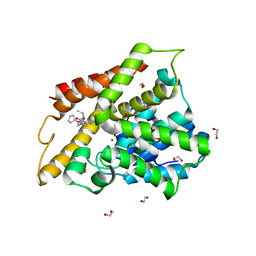 | | Crystal structure of the catalytic domain of PDE4D2 with compound 12c | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL8
 
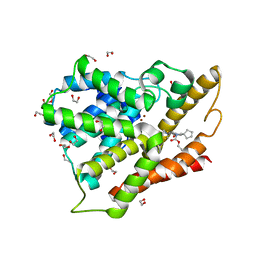 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL3
 
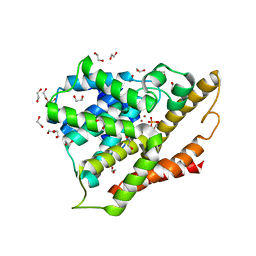 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4TKX
 
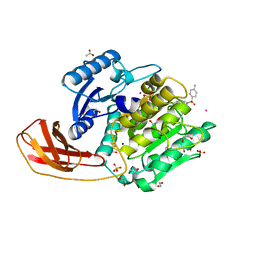 | | Structure of Protease | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, LEAD (II) ION, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2014-05-28 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the lysine specific protease Kgp from Porphyromonas gingivalis, a target for improved oral health.
Protein Sci., 24, 2015
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
1PFO
 
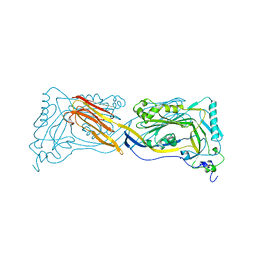 | | PERFRINGOLYSIN O | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form.
Cell(Cambridge,Mass.), 89, 1997
|
|
5IMY
 
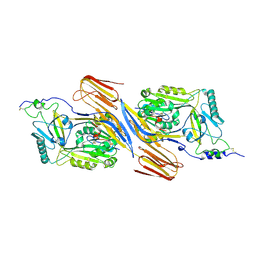 | | Trapped Toxin | | Descriptor: | CD59 glycoprotein, Vaginolysin | | Authors: | Lawrence, S.L, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
2PHM
 
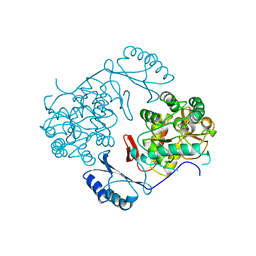 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
1PRE
 
 | | PROAEROLYSIN | | Descriptor: | PROAEROLYSIN | | Authors: | Parker, M.W, Buckley, J.T, Postma, J.P.M, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1995-09-15 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states.
Nature, 367, 1994
|
|
1PHZ
 
 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
