2KB5
 
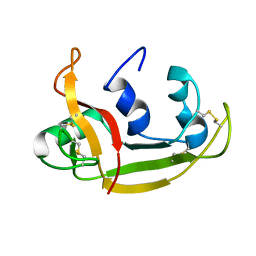 | | Solution NMR Structure of Eosinophil Cationic Protein/RNase 3 | | Descriptor: | Eosinophil cationic protein | | Authors: | Rico, M, Bruix, M, Laurents, D.V, Santoro, J, Jimenez, M, Boix, E, Moussaoui, M, Nogues, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The (1)H, (13)C, (15)N resonance assignment, solution structure, and residue level stability of eosinophil cationic protein/RNase 3 determined by NMR spectroscopy
Biopolymers, 91, 2009
|
|
2JON
 
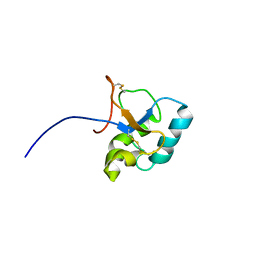 | | Solution structure of the C-terminal domain Ole e 9 | | Descriptor: | Beta-1,3-glucanase | | Authors: | Trevino, M.A, Palomares, O, Castrillo, I, Villalba, M, Rodriguez, R, Rico, M, Santoro, J, Bruix, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ole e 9, a major allergen of olive pollen
Protein Sci., 17, 2008
|
|
2LVZ
 
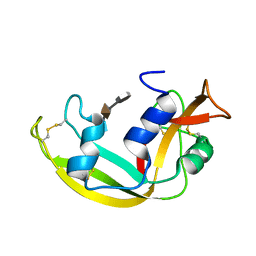 | | Solution structure of a Eosinophil Cationic Protein-trisaccharide heparin mimetic complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-propan-2-yl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, Eosinophil cationic protein | | Authors: | Garcia Mayoral, M, Canales, A, Diaz, D, Lopez Prados, J, Moussaoui, M, de Paz, J, Angulo, J, Nieto, P, Jimenez Barbero, J, Boix, E, Bruix, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Insights into the glycosaminoglycan-mediated cytotoxic mechanism of eosinophil cationic protein revealed by NMR.
Acs Chem.Biol., 8, 2013
|
|
2NCS
 
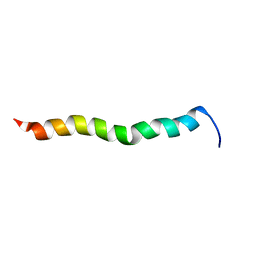 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2NCT
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2LDO
 
 | | Solution structure of triheme cytochrome PpcA from Geobacter sulfurreducens reveals the structural origin of the redox-Bohr effect | | Descriptor: | Cytochrome c3, HEME C | | Authors: | Morgado, L, Paixao, V.B, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | Revealing the structural origin of the redox-Bohr effect: the first solution structure of a cytochrome from Geobacter sulfurreducens.
Biochem.J., 441, 2012
|
|
2K11
 
 | | Solution structure of human pancreatic ribonuclease | | Descriptor: | Pancreatic Ribonuclease | | Authors: | Kover, K.E, Bruix, M, Santoro, J, Batta, G, Laurents, D.V, Rico, M. | | Deposit date: | 2008-02-20 | | Release date: | 2008-06-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of human pancreatic ribonuclease determined by NMR spectroscopy provide insight into its remarkable biological activities and inhibition.
J.Mol.Biol., 379, 2008
|
|
2KAA
 
 | |
2L38
 
 | | R29Q Sticholysin II mutant | | Descriptor: | Sticholysin-2 | | Authors: | Castrillo, I, Alegre-Cebollada, J, Martinez-del-Pozo, A, Gavilanes, J, Bruix, M. | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of StnIIR29Q, a defective lipid binding mutant of the sea anemone actinoporin Sticholysin II
To be Published
|
|
2JMC
 
 | | Chimer between Spc-SH3 and P41 | | Descriptor: | Spectrin alpha chain, brain and P41 peptide chimera | | Authors: | van Nuland, N.A.J, Candel, A.M, Martinez, J.C, Conejero-Lara, F, Bruix, M. | | Deposit date: | 2006-11-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of a single-chain chimeric protein mimicking a SH3-peptide complex
Febs Lett., 581, 2007
|
|
2MAR
 
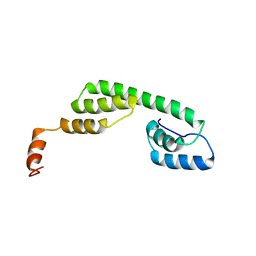 | | Solution structure of Ani s 5 Anisakis simplex allergen | | Descriptor: | SXP/RAL-2 family protein | | Authors: | Garcia-Mayoral, M.F, Trevino, M.A, Perez-Pinar, T, Caballero, M.L, Knaute, T, Umpierrez, A, Bruix, M, Rodriguez-Perez, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Relationships between IgE/IgG4 epitopes, structure and function in Anisakis simplex Ani s 5, a member of the SXP/RAL-2 protein family
Plos Negl Trop Dis, 8, 2014
|
|
2MZ9
 
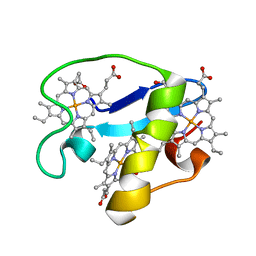 | | Solution structure of oxidized triheme cytochrome PpcA from Geobacter sulfurreducens | | Descriptor: | HEME C, PpcA | | Authors: | Morgado, L, Bruix, M, Pokkuluri, R, Salgueiro, C.A, Turner, D.L. | | Deposit date: | 2015-02-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox- and pH-linked conformational changes in triheme cytochrome PpcA from Geobacter sulfurreducens.
Biochem. J., 474, 2017
|
|
2KS4
 
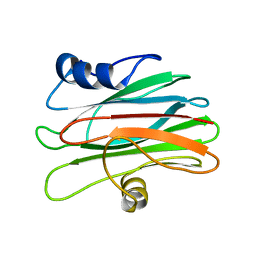 | |
2L2B
 
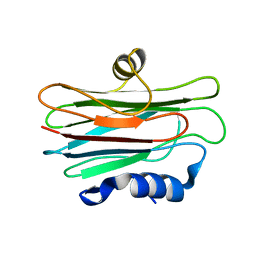 | |
2N91
 
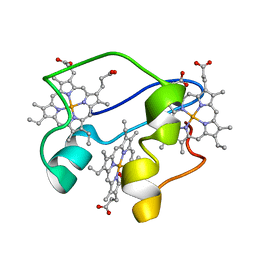 | | A key amino acid in the control of different functional behavior within the triheme cytochrome family from Geobacter sulfurreducens | | Descriptor: | Cytochrome C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dantas, J.M, Simoes, T, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2015-11-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unveiling the Structural Basis That Regulates the Energy Transduction Properties within a Family of Triheme Cytochromes from Geobacter sulfurreducens.
J.Phys.Chem.B, 120, 2016
|
|
2LT5
 
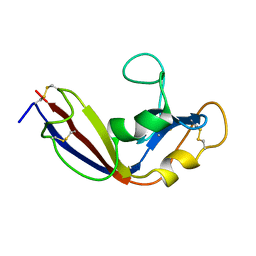 | | Zymogen-FLG of the onconase | | Descriptor: | Protein P-30 | | Authors: | Vilanova, M, Callis, M, Laurents, D.V, Ribo, M, Bruix, M, Serrano, S. | | Deposit date: | 2012-05-14 | | Release date: | 2012-10-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: |
|
|
1T0W
 
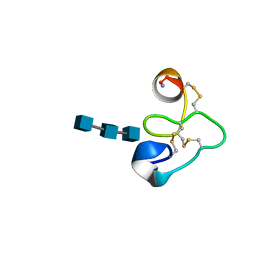 | | 25 NMR structures of Truncated Hevein of 32 aa (Hevein-32) complex with N,N,N-triacetylglucosamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevein | | Authors: | Aboitiz, N, Vila-Perello, M, Groves, P, Asensio, J.L, Andreu, D, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR and modeling studies of protein-carbohydrate interactions: synthesis, three-dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides
Chembiochem, 5, 2004
|
|
4CVD
 
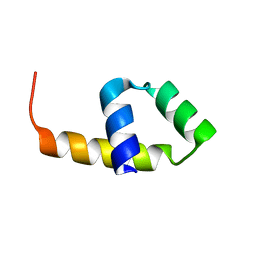 | |
4UHU
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | Descriptor: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
3BXU
 
 | | PpcB, A Cytochrome c7 from Geobacter sulfurreducens | | Descriptor: | Cytochrome c3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2008-01-14 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the modulation of the redox properties of two Geobacter sulfurreducens homologous triheme cytochromes.
Biochim.Biophys.Acta, 1777, 2008
|
|
1TRL
 
 | | NMR SOLUTION STRUCTURE OF THE C-TERMINAL FRAGMENT 255-316 OF THERMOLYSIN: A DIMER FORMED BY SUBUNITS HAVING THE NATIVE STRUCTURE | | Descriptor: | THERMOLYSIN FRAGMENT 255 - 316 | | Authors: | Rico, M, Jimenez, M.A, Gonzalez, C, De Filippis, V, Fontana, A. | | Deposit date: | 1994-09-02 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the C-terminal fragment 255-316 of thermolysin: a dimer formed by subunits having the native structure.
Biochemistry, 33, 1994
|
|
4CNL
 
 | | Crystal structure of the Choline-binding domain of CbpL from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, GLYCEROL, PUTATIVE PNEUMOCOCCAL SURFACE PROTEIN, ... | | Authors: | Gutierrez-Fernandez, J, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Architecture and Unique Teichoic Acid Recognition Features of Choline-Binding Protein L (Cbpl) Contributing to Pneumococcal Pathogenesis.
Sci.Rep., 6, 2016
|
|
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
5MF3
 
 | | NMR solution structure of Harzianin HK-VI in SDS micelles | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin HK-VI: conformational and biological analysis
To Be Published
|
|
5MF8
 
 | | NMR solution structure of Harzianin HK-VI in trifluoroethanol | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin HK-VI: conformational and biological analysis
To Be Published
|
|
