5MWC
 
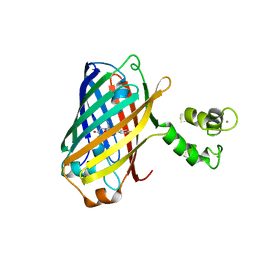 | | Crystal structure of the genetically-encoded green calcium indicator NTnC in its calcium bound state | | Descriptor: | CALCIUM ION, genetically-encoded green calcium indicator NTnC | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Rakitina, T.V, Popov, V.O, Subach, O.M, Barykina, N.V, Subach, F.V. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enchanced variant of genetically-encoded green calcium indicator NTnC
To Be Published
|
|
6Q8E
 
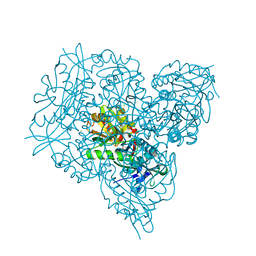 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain-amino-acid aminotransferase, CHLORIDE ION | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
6ET6
 
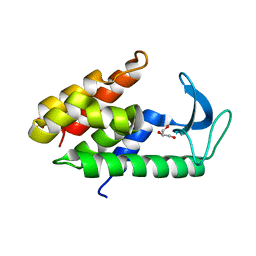 | | Crystal structure of muramidase from Acinetobacter baumannii AB 5075UW prophage | | Descriptor: | GLYCEROL, Lysozyme, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Sykilinda, N.N, Shneider, M.M, Miroshnikov, K.A, Popov, V.O. | | Deposit date: | 2017-10-25 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of anAcinetobacterBroad-Range Prophage Endolysin Reveals a C-Terminal alpha-Helix with the Proposed Role in Activity against Live Bacterial Cells.
Viruses, 10, 2018
|
|
6FUC
 
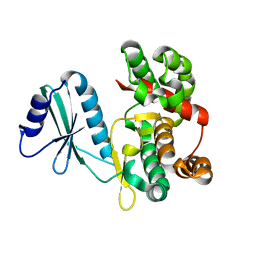 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 | | Descriptor: | Aminoglycoside phosphotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6FUX
 
 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 in complex with ADP and streptomycin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, GLYCEROL, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6GKR
 
 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum in PLP-form (holo-form) | | Descriptor: | ACETATE ION, Branched-chain-amino-acid aminotransferase, CHLORIDE ION, ... | | Authors: | Boyko, K.M, Bezsudnova, E.Y, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum.
Biochimie, 158, 2018
|
|
7PPP
 
 | | Crystal structure of ZAD-domain of ZNF_276 protein from rabbit. | | Descriptor: | ZINC ION, Zinc finger protein 276 | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7QGK
 
 | | The mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer in its red state | | Descriptor: | MAGNESIUM ION, The red form of the mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Gaivoronskii, F.A, Vlaskina, A.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The mRubyFT Protein, Genetically Encoded Blue-to-Red Fluorescent Timer.
Int J Mol Sci, 23, 2022
|
|
8ZBO
 
 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
7POH
 
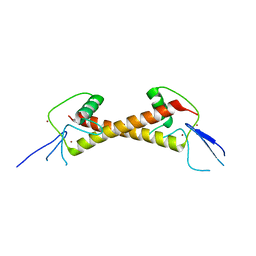 | | Crystal structure of ZAD-domain of Serendipity-d protein from D.melanogaster | | Descriptor: | Serendipity locus protein delta, ZINC ION | | Authors: | Boyko, K.M, Kachalova, G.S, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7POK
 
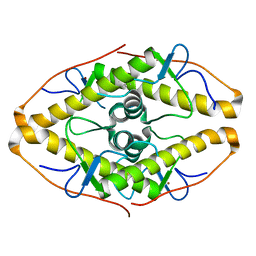 | | Crystal structure of ZAD-domain of Pita protein from D.melanogaster | | Descriptor: | LD15650p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7PO9
 
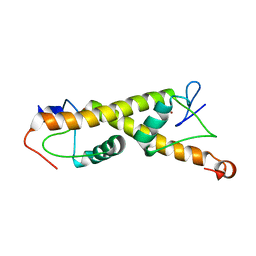 | | Crystal structure of ZAD-domain of M1BP protein from D.melanogaster | | Descriptor: | LD30467p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7OIN
 
 | | Crystal structure of LSSmScarlet - a genetically encoded red fluorescent protein with a large Stokes shift | | Descriptor: | LSSmScarlet - Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift, SODIUM ION, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Dorovatovskii, P.V, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Ivashkina, O.I, Popov, V.O, Subach, F.V. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LSSmScarlet, dCyRFP2s, dCyOFP2s and CRISPRed2s, Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift.
Int J Mol Sci, 22, 2021
|
|
6XW2
 
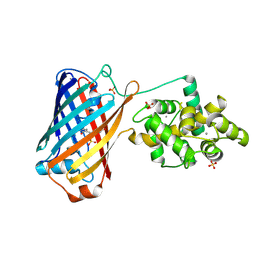 | | Crystal structure of the bright genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein | | Descriptor: | CALCIUM ION, Genetically encoded calcium indicator NCaMP7 based on mNeonGreen fluorescent protein, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Lazarenko, V.A, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein.
Int J Mol Sci, 21, 2020
|
|
6Z1Y
 
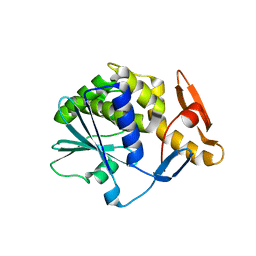 | | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | SODIUM ION, Trichobakin | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Popov, V.O, Usanov, S.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
6XU4
 
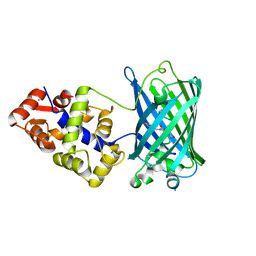 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
7NEA
 
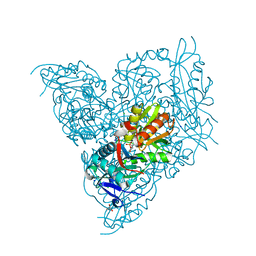 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum (M3 mutant). | | Descriptor: | Branched-chain-amino-acid aminotransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boyko, K.M, Petrova, T, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Suplatov, D.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2021-02-03 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the residues in the active site of the transaminase from Thermobaculum terrenum.
Plos One, 16, 2021
|
|
7NEB
 
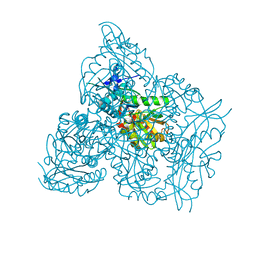 | | Crystal structure of branched-chain amino acid aminotransferase from Thermobaculum terrenum (M4 mutant) | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Boyko, K.M, Petrova, T, Nikolaeva, A.Y, Zeifman, Y.S, Rakitina, T.V, Suplatov, D.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2021-02-03 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the role of the residues in the active site of the transaminase from Thermobaculum terrenum.
Plos One, 16, 2021
|
|
7P7X
 
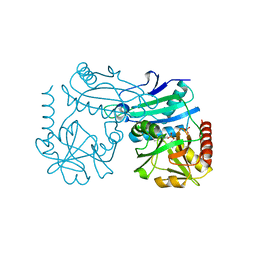 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (holo form). | | Descriptor: | ACETATE ION, Aminotransferase class IV, PHOSPHATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis : Biochemical and Structural Insights into the New Enzyme.
Molecules, 26, 2021
|
|
5CM0
 
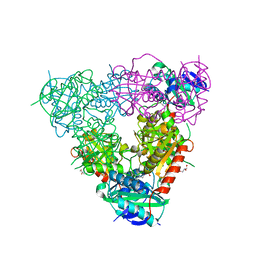 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans | | Descriptor: | Branched-chain transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5E25
 
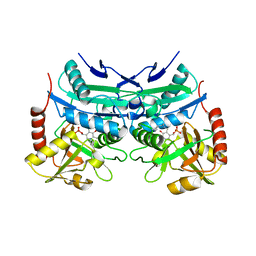 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, branched-chain aminotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
7ARZ
 
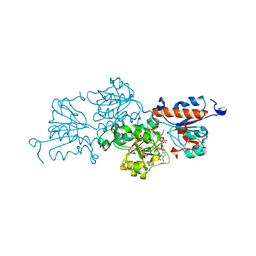 | | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Goryaynova, D.A, Nikolaeva, A.Y, Pometun, A.A, Savin, S.S, Parshin, P.D, Popov, V.O, Tishkov, V.I, Boyko, K.M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens
To Be Published
|
|
8A0H
 
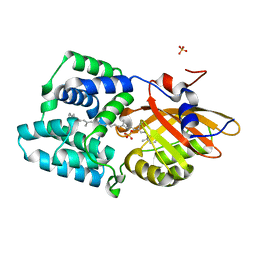 | | Crystal structure of the E25A mutant of the Orange Carotenoid Protein X from Gloeobacter kilaueensis JS1 complexed with echinenone | | Descriptor: | OCP N-terminal domain-containing protein, SULFATE ION, beta,beta-caroten-4-one | | Authors: | Boyko, K.M, Slonimskiy, Y.B, Zupnik, A.O, Varfolomeeva, L.A, Maksimov, E.G, Sluchanko, N.N. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A primordial Orange Carotenoid Protein: Structure, photoswitching activity and evolutionary aspects.
Int.J.Biol.Macromol., 222, 2022
|
|
8AH2
 
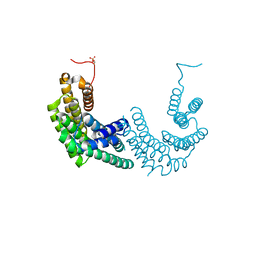 | | Crystal structure of human 14-3-3 zeta fused to the NPM1 peptide including phosphoserine-48 | | Descriptor: | 14-3-3 protein zeta/delta,Nucleophosmin | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Sluchanko, N.N. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the recognition by 14-3-3 proteins of a conditional binding site within the oligomerization domain of human nucleophosmin.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
8P1H
 
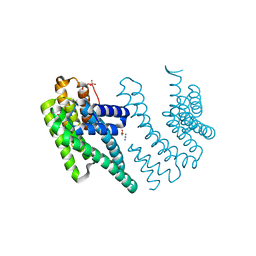 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
