2AHD
 
 | |
1OY5
 
 | | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Liu, J, Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold.
Proteins, 53, 2003
|
|
3QQN
 
 | | The retinal specific CD147 Ig0 domain: from molecular structure to biological activity | | Descriptor: | Basigin | | Authors: | Redzic, J.S, Armstrong, G.S, Isern, N.G, Kieft, J.S, Eisenmesser, E.Z, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-02-15 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | The Retinal Specific CD147 Ig0 Domain: From Molecular Structure to Biological Activity.
J.Mol.Biol., 411, 2011
|
|
3QR2
 
 | | Wild type CD147 Ig0 domain | | Descriptor: | Basigin | | Authors: | Redzic, J.S, Armstrong, G.S, Isern, N.G, Kieft, J.S, Eisenmesser, E.Z, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2011-02-16 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Retinal Specific CD147 Ig0 Domain: From Molecular Structure to Biological Activity.
J.Mol.Biol., 411, 2011
|
|
1LQL
 
 | | Crystal structure of OsmC like protein from Mycoplasma pneumoniae | | Descriptor: | osmotical inducible protein C like family | | Authors: | Choi, I.-G, Shin, D.H, Brandsen, J, Jancarik, J, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-05-10 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of a stress inducible protein from Mycoplasma pneumoniae at 2.85 A resolution
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
3M94
 
 | | Complex crystal structure of Ascaris suum eIF4E-3 with m2,2,7G cap | | Descriptor: | ACETYL GROUP, Eukaryotic translation initiation factor 4E-binding protein 1, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Liu, W, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2010-03-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nematode eIF4E binding an m2,2,7G-Cap and its implications for translation initiation.
Nucleic Acids Res., 39, 2011
|
|
3M93
 
 | |
1LFP
 
 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1XCO
 
 | | Crystal Structure of a Phosphotransacetylase from Bacillus subtilis in complex with acetylphosphate | | Descriptor: | ACETYLPHOSPHATE, Phosphate acetyltransferase, SULFATE ION | | Authors: | Xu, Q.S, Jancarik, J, Lou, Y, Yokota, H, Adams, P, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-09-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of a phosphotransacetylase from Bacillus subtilis and its complex with acetyl phosphate
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1MGP
 
 | | Hypothetical protein TM841 from Thermotoga maritima reveals fatty acid binding function | | Descriptor: | Hypothetical protein TM841, PALMITIC ACID | | Authors: | Schulze-Gahmen, U, Pelaschier, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-08-15 | | Release date: | 2002-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima,
reveals its function as fatty acid binding protein
Proteins, 50, 2003
|
|
2HQB
 
 | |
1ILW
 
 | |
1J97
 
 | | Phospho-Aspartyl Intermediate Analogue of Phosphoserine phosphatase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Cho, H, Wang, W, Kim, R, Yokota, H, Damo, S, Kim, S.-H, Wemmer, D, Kustu, S, Yan, D, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1L7M
 
 | | HIGH RESOLUTION LIGANDED STRUCTURE OF PHOSPHOSERINE PHOSPHATASE (PI COMPLEX) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7P
 
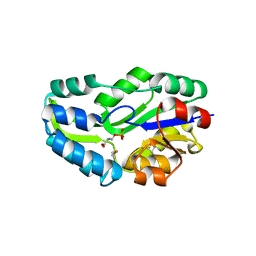 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
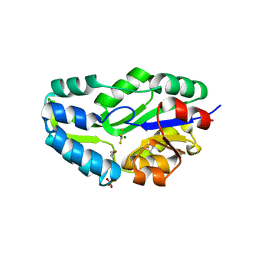 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L2F
 
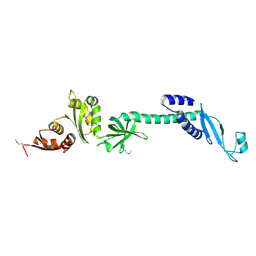 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1L7O
 
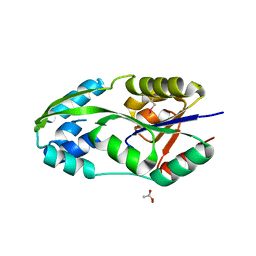 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1JEO
 
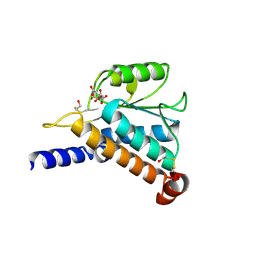 | | Crystal Structure of the Hypothetical Protein MJ1247 from Methanococcus jannaschii at 2.0 A Resolution Infers a Molecular Function of 3-Hexulose-6-Phosphate isomerase. | | Descriptor: | CITRIC ACID, HYPOTHETICAL PROTEIN MJ1247 | | Authors: | Martinez-Cruz, L.A, Dreyer, M.K, Boisvert, D.C, Yokota, H, Martinez-Chantar, M.L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-06-18 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MJ1247 protein from M. jannaschii at 2.0 A resolution infers a molecular function of 3-hexulose-6-phosphate isomerase.
Structure, 10, 2002
|
|
2JMP
 
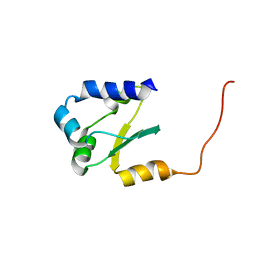 | |
1ZXJ
 
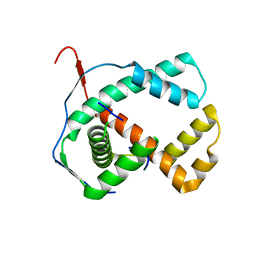 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1TM9
 
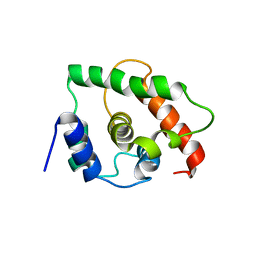 | | NMR Structure of gene target number gi3844938 from Mycoplasma genitalium: Berkeley Structural Genomics Center | | Descriptor: | Hypothetical protein MG354 | | Authors: | Pelton, J.G, Shi, J, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Gene Target gi3844938 from Mycoplasma genitalium
To be Published
|
|
2HEK
 
 | | Crystal structure of O67745, a hypothetical protein from Aquifex aeolicus at 2.0 A resolution. | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oganesyan, V, Jancarik, J, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of O67745_AQUAE, a hypothetical protein from Aquifex aeolicus.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1YTD
 
 | |
1YYX
 
 | |
