5F2V
 
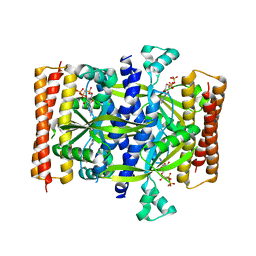 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GTP pyrophosphokinase YjbM, MAGNESIUM ION | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3KTV
 
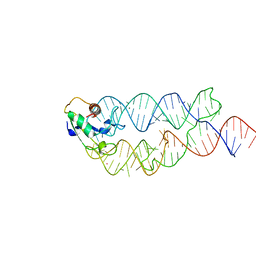 | | Crystal structure of the human SRP19/S-domain SRP RNA complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KTW
 
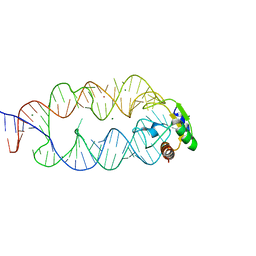 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7BNR
 
 | |
7BNK
 
 | |
4ZOY
 
 | |
4ZOV
 
 | |
4ZOZ
 
 | |
8R9R
 
 | |
5O6U
 
 | | Structure of the Cascade-I-Fv R-loop complex from Shewanella putrefaciens | | Descriptor: | CRISPR-associated protein, Csy4 family, Uncharacterized protein, ... | | Authors: | Pausch, P, Altegoer, F, Bange, G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance.
Mol. Cell, 67, 2017
|
|
5O7H
 
 | | Structure of the Cascade-I-Fv complex from Shewanella putrefaciens | | Descriptor: | CRISPR-associated protein, Csy4 family, Cas5fv, ... | | Authors: | Pausch, P, Altegoer, F, Bange, G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance.
Mol. Cell, 67, 2017
|
|
7OJ1
 
 | | Bacillus subtilis IMPDH in complex with Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Giammarinaro, P.I, Bange, G. | | Deposit date: | 2021-05-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis.
Nat Microbiol, 7, 2022
|
|
4R1J
 
 | | Crystal structure of Arc1p-C | | Descriptor: | GLYCEROL, GU4 nucleic-binding protein 1 | | Authors: | Altegoer, F, Bange, G. | | Deposit date: | 2014-08-06 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Synthetic Adenylation-Domain-Based tRNA-Aminoacylation Catalyst.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RZ2
 
 | |
4RZ3
 
 | | Crystal structure of the MinD-like ATPase FlhG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Site-determining protein | | Authors: | Schuhmacher, J.S, Bange, G. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8A4O
 
 | |
8A14
 
 | |
6H9H
 
 | | Csf5, CRISPR-Cas type IV Cas6 crRNA endonuclease | | Descriptor: | Csf5, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-08-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Nat Microbiol, 4, 2019
|
|
6HJW
 
 | |
6H9I
 
 | | Csf5, CRISPR-Cas type IV Cas6 crRNA endonuclease | | Descriptor: | Csf5, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-08-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Nat Microbiol, 4, 2019
|
|
7O0N
 
 | |
2Q8K
 
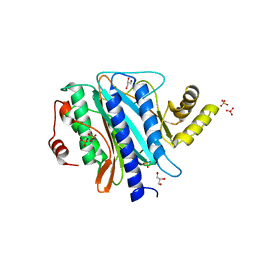 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
2QY9
 
 | |
6YO9
 
 | |
6Z0W
 
 | |
