4ZZ9
 
 | | Crystal structure of T75S mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BNK
 
 | | Crystal structure of T75C mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BMX
 
 | | Crystal structure of T75N mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BRB
 
 | | Crystal structure of Q64E mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-30 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BMW
 
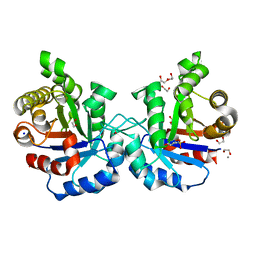 | | Crystal structure of T75V mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5GZP
 
 | |
4YXG
 
 | |
4YWI
 
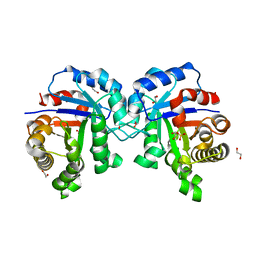 | | F96S/L167V Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-20 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Z0J
 
 | | F96S/S73A Double mutant of Plasmodium Falciparum Triosephosphate Isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Pareek, V, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Connecting Active-Site Loop Conformations and Catalysis in Triosephosphate Isomerase: Insights from a Rare Variation at Residue 96 in the Plasmodial Enzyme
Chembiochem, 17, 2016
|
|
4Z0S
 
 | |
5GV4
 
 | |
2M61
 
 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
1YDV
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Velankar, S.S, Murthy, M.R.N. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design.
Structure, 5, 1997
|
|
5DNI
 
 | | Crystal structure of Methanocaldococcus jannaschii Fumarate hydratase beta subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayaraman, V, Kunala, J, Balaram, H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Revisiting the Burden Borne by Fumarase: Enzymatic Hydration of an Olefin.
Biochemistry, 62, 2023
|
|
7XKY
 
 | |
7D40
 
 | |
7D96
 
 | |
7D97
 
 | |
7D95
 
 | |
1FEV
 
 | |
3BFP
 
 | | Crystal Structure of apo-PglD from Campylobacter jejuni | | Descriptor: | Acetyltransferase, CITRATE ANION | | Authors: | Rangarajan, E.S, Watson, D.C, Leclerc, S, Proteau, A, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-22 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Active Site Residues of PglD, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter jejuni.
Biochemistry, 47, 2008
|
|
2LOC
 
 | | Conotoxin analogue [D-Ala2]BuIIIB | | Descriptor: | Mu-conotoxin BuIIIB | | Authors: | Kuang, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mammalian neuronal sodium channel blocker mu-conotoxin BuIIIB has a structured N-terminus that influences potency.
Acs Chem.Biol., 8, 2013
|
|
2LO9
 
 | | NMR solution structure of Mu-contoxin BuIIIB | | Descriptor: | Mu-conotoxin BuIIIB | | Authors: | Kuang, Z. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mammalian neuronal sodium channel blocker mu-conotoxin BuIIIB has a structured N-terminus that influences potency.
Acs Chem.Biol., 8, 2013
|
|
2M6F
 
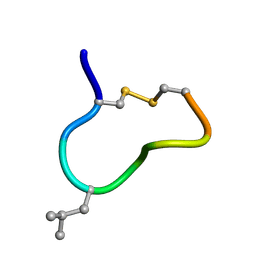 | | NMR solution structure of trans (major) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6H
 
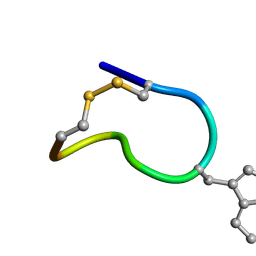 | | Solution structure of trans(C2-P3) trans (D5-P6) of LO959 in methanol | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
