7BON
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7BOM
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr immobilized with gold ions. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7C3B
 
 | | Ferredoxin reductase in carbazole 1,9a-dioxygenase (FAD apo form) | | Descriptor: | ACETATE ION, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ashikawa, Y, Fujimoto, Z, Nojiri, H. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the ferredoxin reductase component of carbazole 1,9a-dioxygenase from Janthinobacterium sp. J3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7C3A
 
 | | Ferredoxin reductase in carbazole 1,9a-dioxygenase | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ashikawa, Y, Fujimoto, Z, Nojiri, H. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the ferredoxin reductase component of carbazole 1,9a-dioxygenase from Janthinobacterium sp. J3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3VGO
 
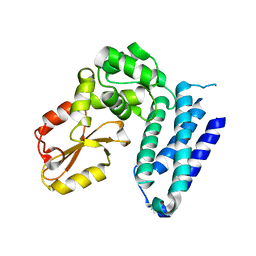 | |
3WVX
 
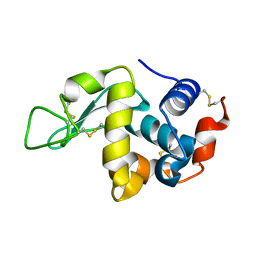 | |
3WUQ
 
 | | Structure of the entire stalk region of the dynein motor domain | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1 | | Authors: | Nishikawa, Y, Oyama, T, Kamiya, N, Kon, T, Toyoshima, Y.Y, Nakamura, H, Kurisu, G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the entire stalk region of the Dynein motor domain
J.Mol.Biol., 426, 2014
|
|
3WVY
 
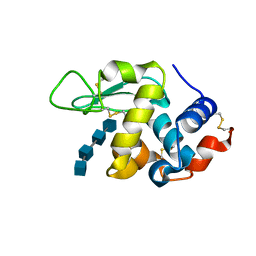 | | Structure of D48A hen egg white lysozyme in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Kawaguchi, Y, Yoneda, K, Araki, T. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The role of Asp48 in the hydrogen bonding network involving Asp52 of hen egg white lysozyme
TO BE PUBLISHED
|
|
3WYH
 
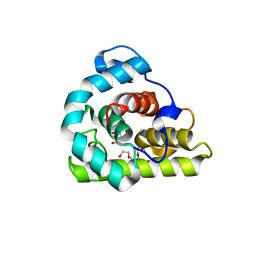 | |
5ZLQ
 
 | | Crystal Structure of C1orf123 | | Descriptor: | UPF0587 protein C1orf123, ZINC ION | | Authors: | Furukawa, Y, Lim, C.T, Tosha, T. | | Deposit date: | 2018-03-29 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a novel zinc-binding protein, C1orf123, as an interactor with a heavy metal-associated domain
PLoS ONE, 13, 2018
|
|
7X1L
 
 | | Malate dehydrogenase from Geobacillus stearothermophilus (gs-MDH) delta E311 mutant complexed with Nicotinamide Adenine Dinucleotide (NAD+) | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Reducing substrate inhibition of malate dehydrogenase from Geobacillus stearothermophilus by C-terminal truncation.
Protein Eng.Des.Sel., 35, 2022
|
|
6K9J
 
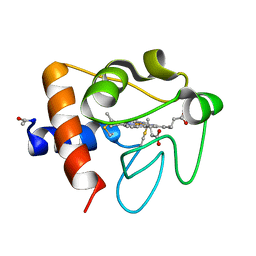 | |
8H8L
 
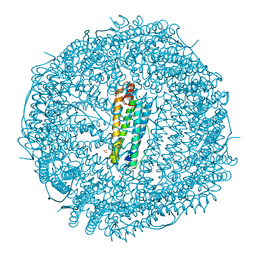 | | Crystal structure of apo-R52F/E56F/R59F/E63F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8M
 
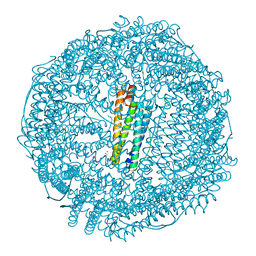 | | Crystal structure of apo-E53F/E57F/E60F/E64F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8O
 
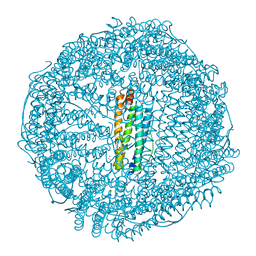 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8N
 
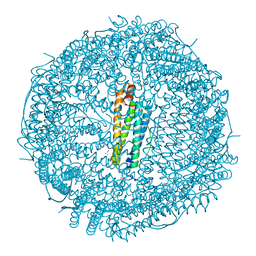 | | Crystal structure of apo-R52Y/E56Y/R59Y/E63Y-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
7VZ4
 
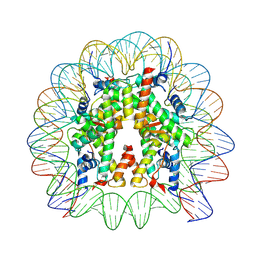 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601L DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Sato, S, Danev, R, Kurumizaka, H. | | Deposit date: | 2021-11-15 | | Release date: | 2023-05-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Methods for High Resolution Cryo-EM Analyses of Nucleosomes
To Be Published
|
|
1ULI
 
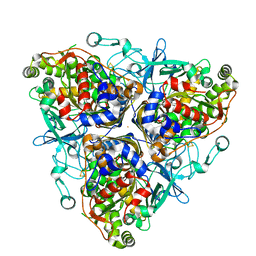 | | Biphenyl dioxygenase (BphA1A2) derived from Rhodococcus sp. strain RHA1 | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, biphenyl dioxygenase large subunit, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
1ULJ
 
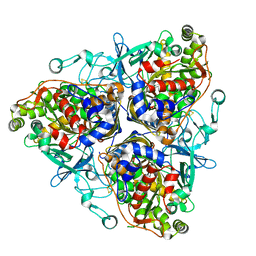 | | Biphenyl dioxygenase (BphA1A2) in complex with the substrate | | Descriptor: | BIPHENYL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
1VDZ
 
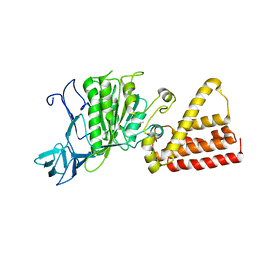 | | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, A-type ATPase subunit A | | Authors: | Maegawa, Y, Morita, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of A-type ATPase catalytic subunit A from Pyrococcus horikoshii OT3
To be Published
|
|
3A65
 
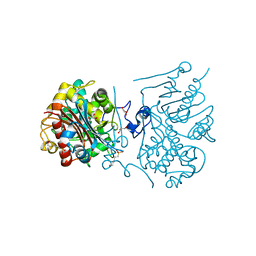 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3A66
 
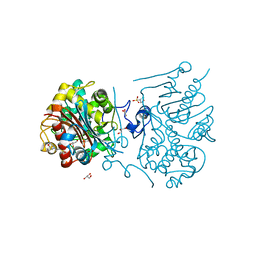 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N/D370Y mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOATE-DIMER HYDROLASE, 6-AMINOHEXANOIC ACID, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
2EYX
 
 | |
2EYZ
 
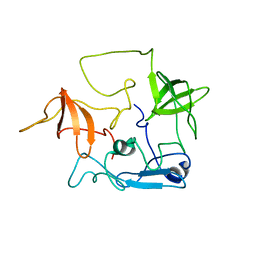 | | CT10-Regulated Kinase isoform II | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYV
 
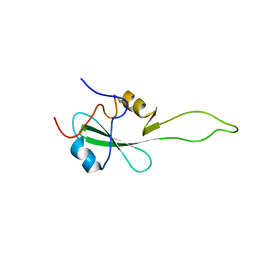 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
