1XD0
 
 | | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, Alpha-amylase, ... | | Authors: | Li, C, Begum, A, Numao, S, Park, K.H, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-03 | | Release date: | 2004-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts
Biochemistry, 44, 2005
|
|
1XH1
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with chloride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, pancreatic, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1JAK
 
 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
1XGZ
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, pancreatic, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1XH0
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED HEXASACCHARIDE, Alpha-amylase, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1XH2
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase complexed with chloride and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED PENTASACCHARIDE, Alpha-amylase, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
1XSI
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETIC ACID, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XSK
 
 | | Structure of a Family 31 alpha glycosidase glycosyl-enzyme intermediate | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 5(R)-fluoro-beta-D-xylopyranose, Putative family 31 glucosidase yicI, ... | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 {alpha}-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XSJ
 
 | | Structure of a Family 31 alpha glycosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative family 31 glucosidase yicI | | Authors: | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2004-10-19 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
1XD1
 
 | | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACARBOSE DERIVED HEXASACCHARIDE, Alpha-amylase, ... | | Authors: | Li, C, Begum, A, Numao, S, Park, K.H, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-03 | | Release date: | 2004-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acarbose Rearrangement Mechanism Implied by the Kinetic and Structural Analysis of Human Pancreatic alpha-Amylase in Complex with Analogues and Their Elongated Counterparts
Biochemistry, 44, 2005
|
|
2A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Sulzenbacher, G, Mackenzie, L, Withers, S.G, Divne, C, Jones, T.A, Woldike, H.F, Schulein, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335, 1998
|
|
2AH2
 
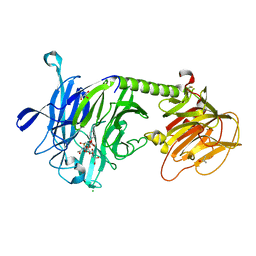 | | Trypanosoma cruzi trans-sialidase in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase
Structure, 12, 2004
|
|
2YK7
 
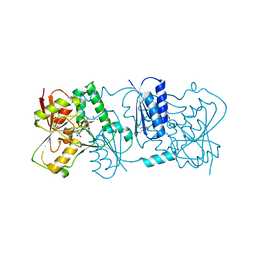 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP-3F-Neu5Ac. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK4
 
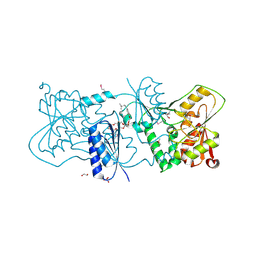 | | Structure of Neisseria LOS-specific sialyltransferase (NST). | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK5
 
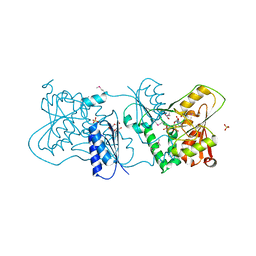 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CMP. | | Descriptor: | 1,2-ETHANEDIOL, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2YK6
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CDP. | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-DIPHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
3D4Z
 
 | | GOLGI MANNOSIDASE II complex with gluco-imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, GLUCOIMIDAZOLE, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3SCR
 
 | | Crystal Structure of Rice BGlu1 E386S Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCN
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCT
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCS
 
 | | Crystal Structure of Rice BGlu1 E386S Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCU
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with Cellopentaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCO
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCV
 
 | | Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCW
 
 | | Crystal Structure of Rice BGlu1 E386G/Y341A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
