1RCO
 
 | | SPINACH RUBISCO IN COMPLEX WITH THE INHIBITOR D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE | | Descriptor: | D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-10-31 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
J.Biol.Chem., 271, 1996
|
|
1RBO
 
 | | SPINACH RUBISCO IN COMPLEX WITH THE INHIBITOR 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-10-31 | | Release date: | 1997-03-12 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
J.Biol.Chem., 271, 1996
|
|
8A5R
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized and measured in dark. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
8A5S
 
 | | Crystal structure of light-activated DNA-binding protein EL222 from Erythrobacter litoralis crystallized in dark, measured illuminated. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Koval, T, Chaudhari, A, Fuertes, G, Andersson, I, Dohnalek, J. | | Deposit date: | 2022-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EL222 from Erythrobacter litoralis.
To Be Published
|
|
5OYA
 
 | | Unusual posttranslational modifications revealed in crystal structures of diatom Rubisco. | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Valegard, K, Andersson, I. | | Deposit date: | 2017-09-08 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications.
J. Biol. Chem., 293, 2018
|
|
2JAH
 
 | | Biochemical and structural analysis of the Clavulanic acid dehydeogenase (CAD) from Streptomyces clavuligerus | | Descriptor: | CLAVULANIC ACID DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
2JB8
 
 | | Deacetoxycephalosporin C synthase complexed with 5-hydroxy-4-keto valeric acid | | Descriptor: | 4,5-DIOXOPENTANOIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (III) ION | | Authors: | Cicero, G, Carbonera, C, Valegard, K, Hajdu, J, Andersson, I, Ranghino, G. | | Deposit date: | 2006-12-05 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Study of the Oxidative Half-Reaction Catalysed by a Non-Heme Ferrous Catalytic Centre by Means of Structural and Computational Methodologies
Int.J.Quantum Chem., 107, 2007
|
|
2JAP
 
 | | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the beta-Lactamase Inhibitor Clavulanic acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, CLAVALDEHYDE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | MacKenzie, A.K, Kershaw, N.J, Hernandez, H, Robinson, C.V, Schofield, C.J, Andersson, I. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clavulanic Acid Dehydrogenase: Structural and Biochemical Analysis of the Final Step in the Biosynthesis of the Beta-Lactamase Inhibitor Clavulanic Acid
Biochemistry, 46, 2007
|
|
6RTS
 
 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus complexed with NAD+ | | Descriptor: | ACETATE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RTT
 
 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus complexed with picolinic acid | | Descriptor: | GLYCEROL, PYRIDINE-2-CARBOXYLIC ACID, SULFATE ION, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RTU
 
 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus complexed with alpha-aminoadipic acid | | Descriptor: | 2-AMINOHEXANEDIOIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RTR
 
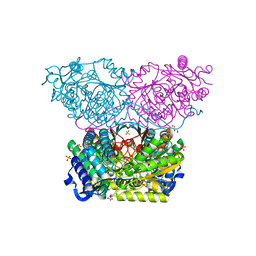 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5LT5
 
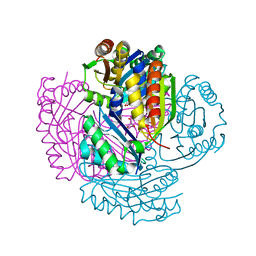 | | Carboxysome shell protein CcmP from Synechococcus elongatus PCC 7942 | | Descriptor: | CHLORIDE ION, CcmP, GLYCEROL | | Authors: | Larsson, A.M, Hasse, D, Valegard, K, Andersson, I. | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore.
J. Exp. Bot., 68, 2017
|
|
5MAC
 
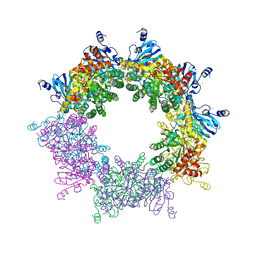 | | Crystal structure of decameric Methanococcoides burtonii Rubisco complexed with 2-carboxyarabinitol bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gunn, L.H, Valegard, K, Andersson, I. | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic.
J. Biol. Chem., 292, 2017
|
|
5LSR
 
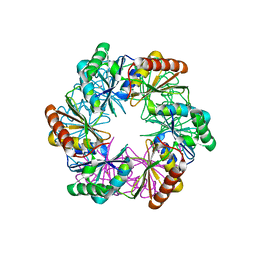 | | Carboxysome shell protein CcmP from Synechococcus elongatus PCC 7942 | | Descriptor: | CcmP, THIOCYANATE ION | | Authors: | Larsson, A.M, Hasse, D, Valegard, K, Andersson, I. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore.
J. Exp. Bot., 68, 2017
|
|
1W2O
 
 | | Deacetoxycephalosporin C synthase (with a N-terminal his-tag) in complex with Fe(II) and deacetoxycephalosporin C | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, DEACETOXYCEPHALOSPORIN-C, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1W2A
 
 | | Deacetoxycephalosporin C synthase (with his-tag) complexed with Fe(II) and ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1W2N
 
 | | Deacetoxycephalosporin C synthase (with a N-terminal his-tag) in complex with Fe(II) and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1W28
 
 | | Conformational flexibility of the C-terminus with implications for substrate binding and catalysis in a new crystal form of deacetoxycephalosporin C synthase | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1W7W
 
 | |
1UO9
 
 | | Deacetoxycephalosporin C synthase complexed with succinate | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, SUCCINIC ACID | | Authors: | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UOB
 
 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and penicillin G | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UWA
 
 | | L290F mutant rubisco from chlamydomonas | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-02-03 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Altered Intersubunit Interactions in Crystal Structures of Catalytically Compromised Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
1UNB
 
 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-09 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UOG
 
 | | Deacetoxycephalosporin C synthase complexed with deacetoxycephalosporin C | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, DEACETOXYCEPHALOSPORIN-C, FE (II) ION | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-17 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
