1KYS
 
 | | Crystal Structure of a Zn-bound Green Fluorescent Protein Biosensor | | Descriptor: | Green Fluorescent Protein, ZINC ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
4CH8
 
 | | High-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
1L4Z
 
 | | X-RAY CRYSTAL STRUCTURE OF THE COMPLEX OF MICROPLASMINOGEN WITH ALPHA DOMAIN OF STREPTOKINASE IN THE PRESENCE CADMIUM IONS | | Descriptor: | CADMIUM ION, Plasminogen, Streptokinase | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-06 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation.
PROTEIN ENG., 15, 2002
|
|
4DBE
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus complexed with inhibitor BMP
To be Published
|
|
4DX3
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue | | Descriptor: | CHLORIDE ION, GLYCEROL, Mandelate racemase / muconate lactonizing enzyme family protein | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue
to be published
|
|
1LGF
 
 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-15 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
1LRO
 
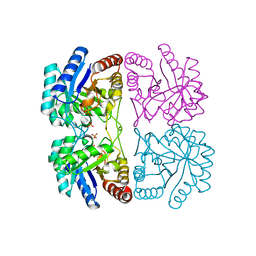 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP and Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1L2Q
 
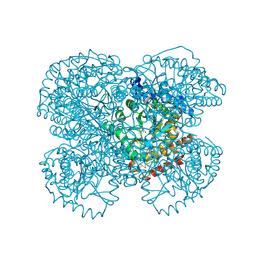 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
4DWO
 
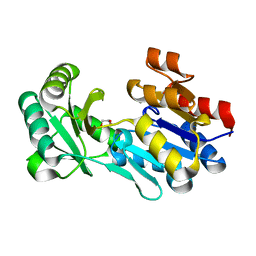 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-26 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II
To be Published
|
|
4DLF
 
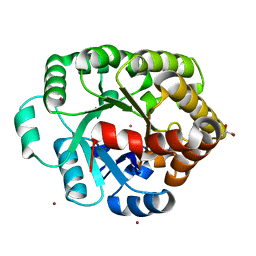 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P3221 | | Descriptor: | Amidohydrolase 2, GLYCEROL, UNKNOWN ATOM OR ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (target efi-500235) with bound ZN, space group P3221
to be published
|
|
4DF7
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23L/V99I at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DFD
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from bacteroides thetaiotaomicron, magnesium complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative haloacid dehalogenase-like hydrolase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4DY0
 
 | | Crystal structure of native protease nexin-1 with heparin | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, GLYCEROL, Glia-derived nexin, ... | | Authors: | Huntington, J.A, Li, W. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of protease nexin-1 in complex with heparin and thrombin suggest a 2-step recognition mechanism.
Blood, 120, 2012
|
|
1L4D
 
 | | CRYSTAL STRUCTURE OF MICROPLASMINOGEN-STREPTOKINASE ALPHA DOMAIN COMPLEX | | Descriptor: | PLASMINOGEN, STREPTOKINASE, SULFATE ION | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation
PROTEIN ENG., 15, 2002
|
|
4DX6
 
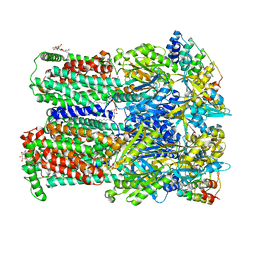 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DODECYL-BETA-D-MALTOSIDE | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DEY
 
 | |
4DGQ
 
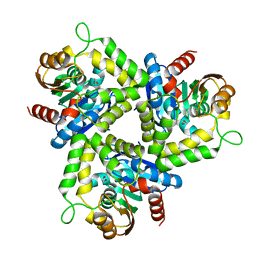 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
1L98
 
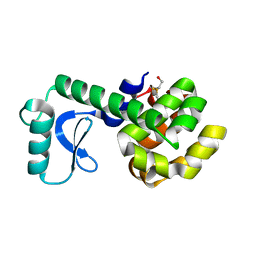 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1LYE
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
4EET
 
 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4ENN
 
 | |
1LYG
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
1LYI
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
4EEP
 
 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EK3
 
 | |
