8UKM
 
 | | T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml30-redesigned-4-OT-fold, T33-ml30-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
6VGS
 
 | | Alpha-ketoisovalerate decarboxylase (KivD) from Lactococcus lactis, thermostable mutant | | Descriptor: | Alpha-keto acid decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Chan, S, Korman, T.P, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2020-01-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isobutanol production freed from biological limits using synthetic biochemistry.
Nat Commun, 11, 2020
|
|
3HYD
 
 | |
3I6P
 
 | |
3IA0
 
 | |
3I96
 
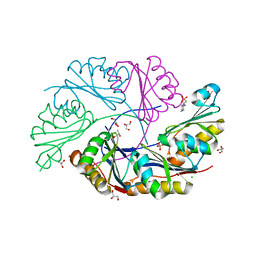 | | Ethanolamine Utilization Microcompartment Shell Subunit, EutS | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Ethanolamine utilization protein eutS, ... | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2009-07-10 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanisms of a Protein-Based Organelle in Escherichia coli.
Science, 327, 2010
|
|
3I87
 
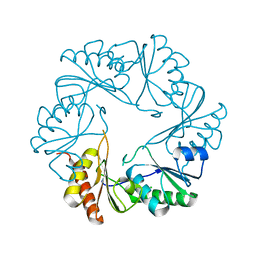 | |
3I71
 
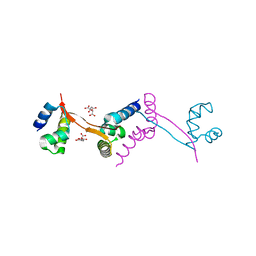 | |
3I82
 
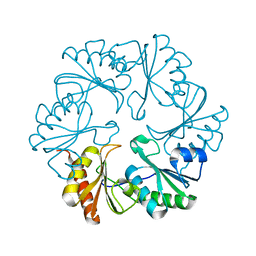 | |
3K6V
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Citrate-Bound Open Form | | Descriptor: | CITRIC ACID, Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6W
 
 | | Apo and ligand bound structures of ModA from the archaeon Methanosarcina acetivorans | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6X
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Molybdate-Bound Close Form with 2 Molecules in Asymmetric Unit Forming Beta Barrel | | Descriptor: | MOLYBDATE ION, SULFATE ION, Solute-binding protein MA_0280 | | Authors: | Chan, S, Chernishof, I, Giuroiu, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3K6U
 
 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3L1F
 
 | | Bovine AlphaA crystallin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-crystallin A chain | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3L1E
 
 | | Bovine AlphaA crystallin Zinc Bound | | Descriptor: | Alpha-crystallin A chain, GLYCEROL, ZINC ION | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3L1G
 
 | | Human AlphaB crystallin | | Descriptor: | Alpha-crystallin B chain, SULFATE ION | | Authors: | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2009-12-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3FPO
 
 | |
3CIM
 
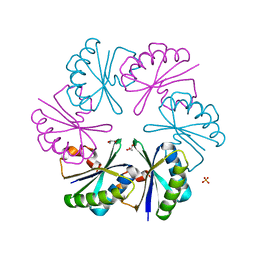 | | Carboxysome shell protein, CcmK2 C-terminal deletion mutant | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 2, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2008-03-11 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Insights from multiple structures of the shell proteins from the beta-carboxysome.
Protein Sci., 18, 2009
|
|
3CGI
 
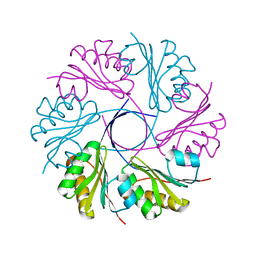 | |
1YJO
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
1YJP
 
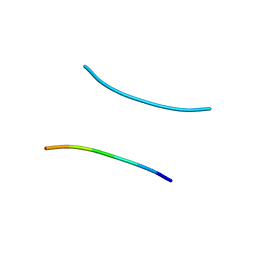 | | Structure of GNNQQNY from yeast prion Sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
1YKW
 
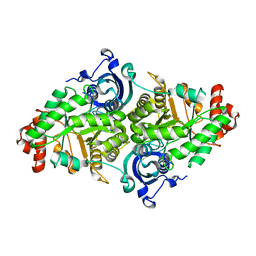 | |
3FOD
 
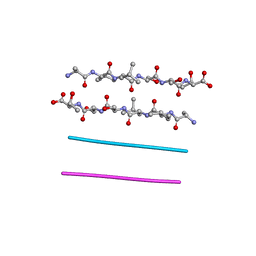 | |
3DGJ
 
 | | NNFGAIL segment from Islet Amyloid Polypeptide (IAPP or amylin) | | Descriptor: | NNFGAIL peptide | | Authors: | Wiltzius, J.J, Sievers, S.A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2008-06-13 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin).
Protein Sci., 17, 2008
|
|
1Z8F
 
 | | Guanylate Kinase Double Mutant A58C, T157C from Mycobacterium tuberculosis (Rv1389) | | Descriptor: | FORMIC ACID, Guanylate kinase | | Authors: | Chan, S, Sawaya, M.R, Choi, B, Zocchi, G, Perry, L.J, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-03-30 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Guanylate Kinase from Mycobacterium tuberculosis (Rv1389)
To be Published
|
|
