8W0M
 
 | |
8W0L
 
 | |
8UZK
 
 | |
8VJ3
 
 | |
8VQZ
 
 | |
8VQW
 
 | |
8VJ2
 
 | |
8W21
 
 | |
8W0D
 
 | |
8UR2
 
 | |
9AZZ
 
 | |
8W0B
 
 | |
8VR0
 
 | |
8W0C
 
 | |
8VR1
 
 | |
9BKW
 
 | |
8UZN
 
 | |
8SBV
 
 | |
8SBW
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, orthorhombic form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, ACETATE ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, orthorhombic form)
To be published
|
|
8SBY
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (NAD and sulfate bound, hexagonal form)
To be published
|
|
8SBX
 
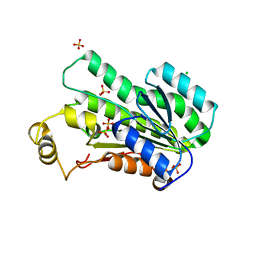 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, hexagonal form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, hexagonal form)
To be published
|
|
8U9A
 
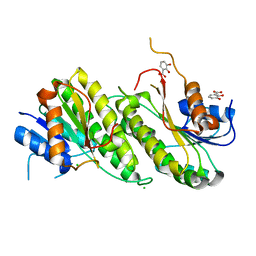 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (DBH bound) | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-2,3-dehydrogenase, CHLORIDE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-09-18 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (DBH bound)
To be published
|
|
8EES
 
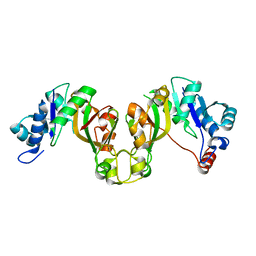 | |
8F8U
 
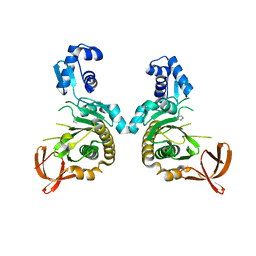 | |
8FI3
 
 | |
