9K0A
 
 | | Structure of Pictet-Spenglerases KslB in complex with product | | Descriptor: | (1~{S},3~{S})-1-(3-hydroxy-3-oxopropyl)-2,3,4,9-tetrahydropyrido[3,4-b]indole-1,3-dicarboxylic acid, Cucumopine synthase C-terminal helical bundle domain-containing protein, GLYCEROL | | Authors: | Mori, T, Renata, H, Abe, I. | | Deposit date: | 2024-10-15 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of Pictet-Spenglerases KslB in complex with product
To Be Published
|
|
9NA4
 
 | | IRAK4 in Complex with Compound 18 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl {[(1S,3s)-3-{[(2P)-2-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-5-{[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]carbamoyl}pyridin-4-yl]amino}cyclobutyl]methyl}carbamate | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
9NA6
 
 | | IRAK4 in Complex with Compound 34 | | Descriptor: | (6P)-4-{[(1S)-1-cyanoethyl]amino}-6-[(8S)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
9ONO
 
 | | Fe-bound B. pseudomallei rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9NRC
 
 | | TMPRSS6 in complex with REGN7999 Fab and REGN8023 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2025-03-14 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | A TMPRSS6-inhibiting mAb improves disease in a beta-thalassemia mouse model and reduces iron in healthy humans.
JCI Insight, 10, 2025
|
|
9ONN
 
 | | Co-bound B. pseudomallei Rubrerythrin | | Descriptor: | COBALT (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONM
 
 | | near-apo rubrerythrin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONR
 
 | | rubrerythrin- E53A E124A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Rubrerythrin, THIOCYANATE ION | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9QFL
 
 | | Crystal structure of YTHDF2 in complex with compound 15 (AI-DF2-68) | | Descriptor: | 6-sulfanylidene-1,7-dihydropyrazolo[3,4-d]pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-11 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9ONQ
 
 | | Mn-bound B. pseudomallei rubrerythrin | | Descriptor: | MANGANESE (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9NND
 
 | | Structure of the HERV-K (HML-2) spike complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaked, R, Katz, M, Diskin, R. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | The prefusion structure of the HERV-K (HML-2) Env spike complex.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9QMP
 
 | | E.coli seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase | | Authors: | Cusack, S. | | Deposit date: | 2025-03-24 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A.
Nature, 347, 1990
|
|
9NYO
 
 | |
9R7J
 
 | |
9R2R
 
 | |
9R2V
 
 | | De novo designed M16 protein fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-05-01 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9R2O
 
 | | De novo designed N5 protein fold | | Descriptor: | N5, POTASSIUM ION, THIOCYANATE ION | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9QEL
 
 | |
9QEM
 
 | | Crystal structure of YTHDF2 in complex with compound 3 (AI-DF2-11) | | Descriptor: | 5-[[(3-methoxyphenyl)amino]methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-10 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9NA5
 
 | | IRAK4 in Complex with Compound 24 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-4-({(1s,4S)-4-[1-(difluoromethyl)-1H-pyrazol-4-yl]cyclohexyl}amino)-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 68, 2025
|
|
8DSS
 
 | |
8DTA
 
 | |
8ER7
 
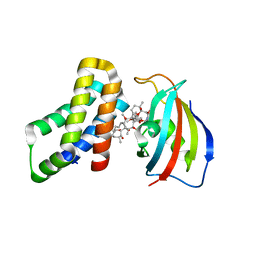 | | FKBP12-FRB in Complex with Compound 12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-9,27-dihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-5,10,21-trimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
8EGK
 
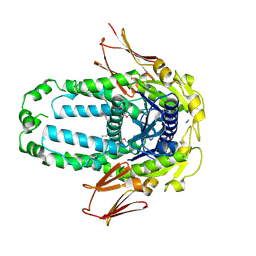 | | Re-refinement of Crystal Structure of NosGet3d, the All4481 protein from Nostoc sp. PCC 7120 | | Descriptor: | AZIDE ION, All4481 protein | | Authors: | Barlow, A.N, Manu, M.S, Ramasamy, S, Clemons Jr, W.M. | | Deposit date: | 2022-09-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
8EB9
 
 | |
