1NLS
 
 | | CONCANAVALIN A AND ITS BOUND SOLVENT AT 0.94A RESOLUTION | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Deacon, A.M, Gleichmann, T, Helliwell, J.R, Kalb(Gilboa), A.J. | | Deposit date: | 1997-01-28 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Structure of Concanavalin a and its Bound Solvent Determined with Small-Molecule Accuracy at 0.94 A Resolution
J.Chem.Soc.,Faraday Trans., 93, 1997
|
|
1CEC
 
 | |
1DEK
 
 | |
1PZC
 
 | | APO-PSEUDOAZURIN (METAL FREE PROTEIN) | | Descriptor: | PSEUDOAZURIN | | Authors: | Petratos, K. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of apo-pseudoazurin from Alcaligenes faecalis S-6.
Febs Lett., 368, 1995
|
|
1CTJ
 
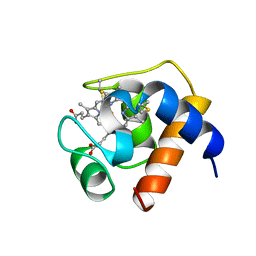 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sheldrick, G.M. | | Deposit date: | 1995-08-08 | | Release date: | 1996-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ab initio determination of the crystal structure of cytochrome c6 and comparison with plastocyanin.
Structure, 3, 1995
|
|
1DEL
 
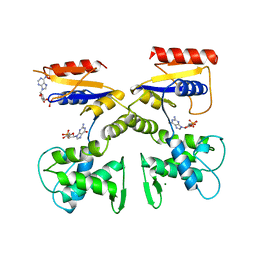 | |
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1GK9
 
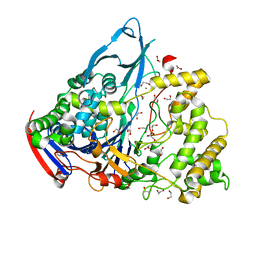 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1H56
 
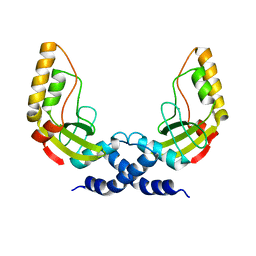 | | Structural and biochemical characterization of a new magnesium ion binding site near Tyr94 in the restriction endonuclease PvuII | | Descriptor: | MAGNESIUM ION, TYPE II RESTRICTION ENZYME PVUII | | Authors: | Spyrida, A, Matzen, C, Lanio, T, Jeltsch, A, Simoncsits, A, Athanasiadis, A, Scheuring-Vanamee, E, Kokkinidis, M, Pingoud, A. | | Deposit date: | 2001-05-20 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Biochemical Characterization of a New Mg(2+) Binding Site Near Tyr94 in the Restriction Endonuclease PvuII.
J.Mol.Biol., 331, 2003
|
|
1H2G
 
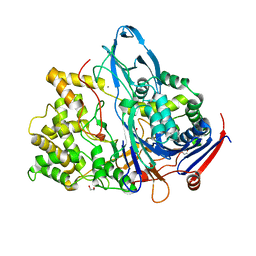 | | Altered substrate specificity mutant of penicillin acylase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Morillas, M, Brannigan, J.A, Ladurner, A.G, Forney, L.J, Virden, R. | | Deposit date: | 2002-08-08 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of Penicillin Acylase Residue B71 Extend Substrate Specificity by Decreasing Steric Constraints for Substrate Binding
Biochem.J., 371, 2003
|
|
1HEU
 
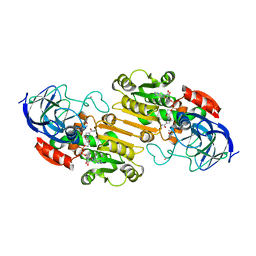 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HF3
 
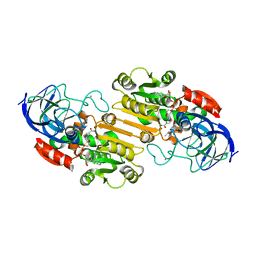 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HF4
 
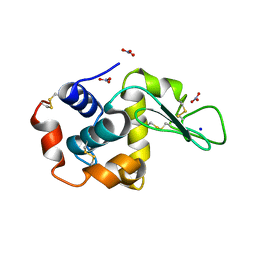 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HSX
 
 | |
1HSW
 
 | |
1RFE
 
 | | Crystal structure of conserved hypothetical protein Rv2991 from Mycobacterium tuberculosis | | Descriptor: | hypothetical protein Rv2991 | | Authors: | Benini, S, Haouz, A, Proux, F, Betton, J.M, Alzari, P, Dodson, G.G, Wilson, K.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-08 | | Release date: | 2004-12-28 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Rv2991 from Mycobacterium tuberculosis: An F420binding protein with unknown function.
J. Struct. Biol., 2019
|
|
5W19
 
 | | Tryptophan indole-lyase complex with oxindolyl-L-alanine | | Descriptor: | 1-carboxy-1-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]azaniumyl}-2-[(3R)-2-oxo-2,3-dihydro-1H-indol-3-yl]ethan-1-ide, POTASSIUM ION, Tryptophanase | | Authors: | Phillips, R.S, Wood, Z.A. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Proteus vulgaris tryptophan indole-lyase complexed with oxindolyl-L-alanine: implications for the reaction mechanism.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1SBN
 
 | |
1RVE
 
 | |
1RPO
 
 | |
1SIB
 
 | |
1IGC
 
 | |
1K3H
 
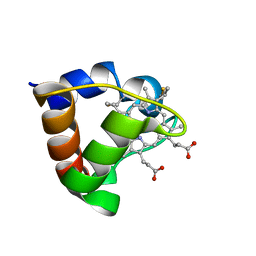 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3G
 
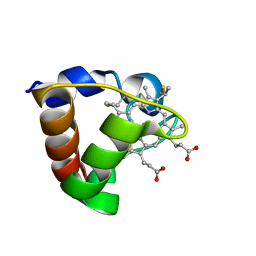 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
