5T9T
 
 | | Protocadherin Gamma B2 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin gamma B2-alpha C, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZQ
 
 | | Protocadherin Gamma A4 extracellular cadherin domains 3-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZN
 
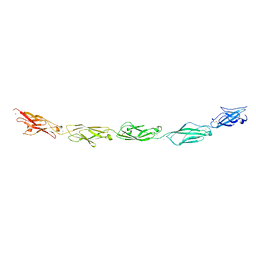 | | Protocadherin gamma A9 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZP
 
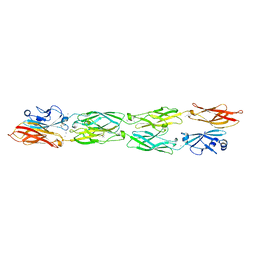 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P21 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin Gamma B7, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
6TA8
 
 | |
3PGT
 
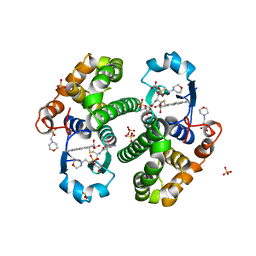 | | CRYSTAL STRUCTURE OF HGSTP1-1[I104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Xiao, B. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
5SZO
 
 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P41212 crystal form | | Descriptor: | CALCIUM ION, Protocadherin Gamma B7, alpha-D-mannopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.612 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5V5X
 
 | | Protocadherin gammaB7 EC3-6 cis-dimer structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protocadherin cis-dimer architecture and recognition unit diversity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DZY
 
 | | Protocadherin beta 8 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Pcdhb8 protein, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5SZM
 
 | | Protocadherin gamma A8 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PcdhgA8 or protocadherin gamma A8, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5SZL
 
 | | Protocadherin gamma A1 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
4HWN
 
 | | Crystal structure of the second Ig-C2 domain of human Fc-receptor like A (FCRLA), Isoform 9 [NYSGRC-005836] | | Descriptor: | Fc receptor-like A | | Authors: | Kumar, P.R, Ahmed, M, Bhosle, R, Calarese, D, Celikigil, A, Chan, M.K, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of the second Ig-C2 domain of the human Fc-receptor like A, Isoform 9 [NYSGRC-005836]
to be published
|
|
1AH2
 
 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | Descriptor: | SERINE PROTEASE PB92 | | Authors: | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
4I9J
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to diC4PC | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9T
 
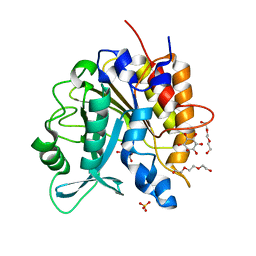 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
8GSS
 
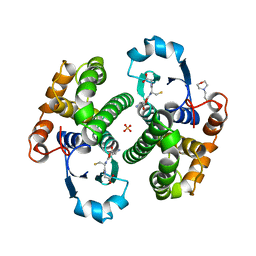 | | Human glutathione S-transferase P1-1, complex with glutathione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
1ASO
 
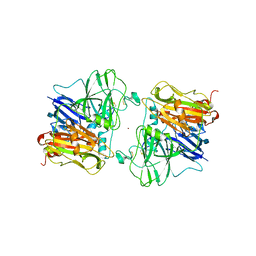 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASQ
 
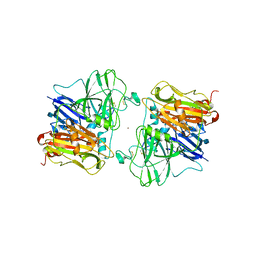 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
4I8Y
 
 | | Structure of the unliganded N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I90
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to choline | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9M
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus bound to HEPES | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
8G5C
 
 | |
8GHA
 
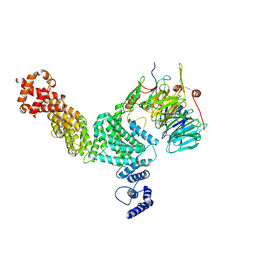 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
8GHM
 
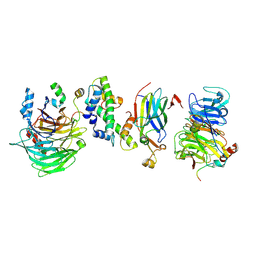 | | Hir1 WD40 domains and Asf1/H3/H4 | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
