8CWG
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWC
 
 | | 20ns Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW7
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW6
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW3
 
 | | 20us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWB
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
6G40
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH9525 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
7AMF
 
 | |
7AMB
 
 | |
5TTC
 
 | | XFEL structure of influenza A M2 wild type TM domain at high pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-11-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6NA5
 
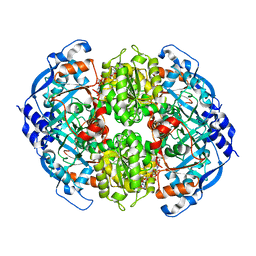 | | Crystal Structure of ECR in complex with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative crotonyl-CoA reductase | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
4YOP
 
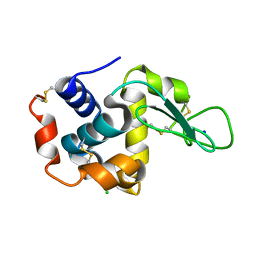 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Nango, E. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Native sulfur/chlorine SAD phasing for serial femtosecond crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6NA4
 
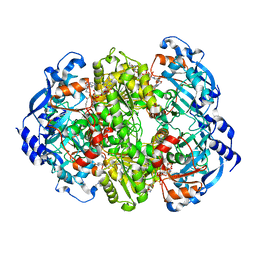 | | Co crystal structure of ECR with Butryl-CoA | | Descriptor: | 9-ETHYL-9H-PURIN-6-YLAMINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
6NA3
 
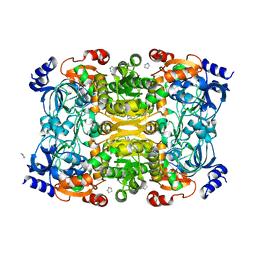 | | Crystal Structure of Apo-form of ECR | | Descriptor: | CHLORIDE ION, Putative crotonyl-CoA reductase, Pyrrolidine | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
6NA6
 
 | |
7AYZ
 
 | |
7AZ0
 
 | |
7AYY
 
 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7JRI
 
 | |
9F9F
 
 | | Laser excitation effects on BR: Extrapolated 6 ps Light dataset recorded at 2493 GW/cm2 at SwissFEL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Bertrand, Q, Weinert, T, Standfuss, J. | | Deposit date: | 2024-05-07 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural effects of high laser power densities on an early bacteriorhodopsin photocycle intermediate.
Nat Commun, 15, 2024
|
|
9F9B
 
 | | Laser excitation effects on BR: reprocessed dark dataset recorded from Nango et al. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Bertrand, Q, Weinert, T, Standfuss, J. | | Deposit date: | 2024-05-07 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural effects of high laser power densities on an early bacteriorhodopsin photocycle intermediate.
Nat Commun, 15, 2024
|
|
9F9I
 
 | | Laser excitation effects on BR: Dark dataset recorded at SACLA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Bertrand, Q, Weinert, T, Standfuss, J. | | Deposit date: | 2024-05-07 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of high laser power densities on an early bacteriorhodopsin photocycle intermediate.
Nat Commun, 15, 2024
|
|
9F9J
 
 | | Laser excitation effects on BR: Extrapolated 10ps Light dataset recorded at 1281 GW/cm2 at SACLA (+100ps, 1ns, 10ns) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Bertrand, Q, Weinert, T, Standfuss, J. | | Deposit date: | 2024-05-07 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural effects of high laser power densities on an early bacteriorhodopsin photocycle intermediate.
Nat Commun, 15, 2024
|
|
9F9H
 
 | | Laser excitation effects on BR: Reprocessed Extrapolated 10ps Light dataset recorded at 525 GW/cm2 from Nogly et al. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Bertrand, Q, Weinert, T, Standfuss, J. | | Deposit date: | 2024-05-07 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural effects of high laser power densities on an early bacteriorhodopsin photocycle intermediate.
Nat Commun, 15, 2024
|
|
