7RW5
 
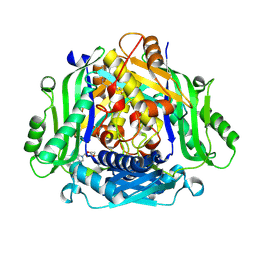 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | Descriptor: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
6M1K
 
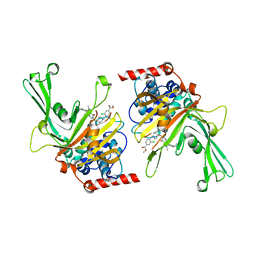 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
6LN0
 
 | |
3V4J
 
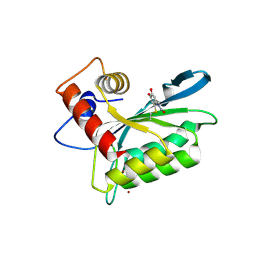 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | 4-[methyl(nitroso)amino]benzene-1,2-diol, DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
3V4K
 
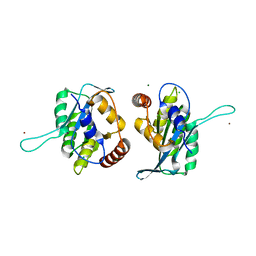 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | CHLORIDE ION, DNA dC->dU-editing enzyme APOBEC-3G, MAGNESIUM ION, ... | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
6AH5
 
 | | Structure of human P2X3 receptor in complex with ATP and Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.819 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
6AH4
 
 | | Structure of human P2X3 receptor in complex with ATP and Ca2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
3IUR
 
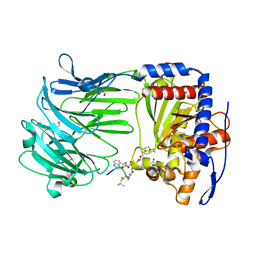 | | apPEP_D266Nx+H2H3 opened state | | Descriptor: | GLYCEROL, H2H3 helices from villin headpiece subdomain HP35, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IVM
 
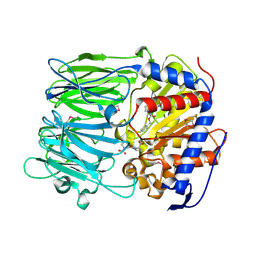 | | apPEP_WT+PP closed state | | Descriptor: | N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-09-01 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUQ
 
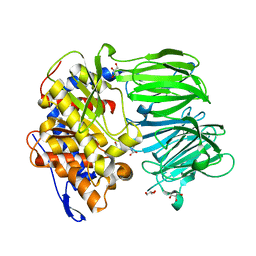 | | apPEP_D622N+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUL
 
 | | apPEP_WT1 opened state | | Descriptor: | GLYCEROL, Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUJ
 
 | | apPEP_WT2 opened state | | Descriptor: | Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUN
 
 | | apPEP_D622N opened state | | Descriptor: | GLYCEROL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUM
 
 | | apPEP_WTX opened state | | Descriptor: | GLYCEROL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
5H5X
 
 | |
4LLF
 
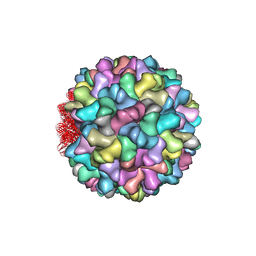 | | Crystal structure of Cucumber Necrosis Virus | | Descriptor: | CALCIUM ION, Capsid protein, ZINC ION | | Authors: | Smith, T. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8891 Å) | | Cite: | Atomic structure of cucumber necrosis virus and the role of the capsid in vector transmission.
J.Virol., 87, 2013
|
|
7WA9
 
 | |
7DG0
 
 | |
7XCZ
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | Authors: | Liu, Z, Lui, S, Gao, Y. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDA
 
 | |
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
6L10
 
 | | PHF20L1 Tudor1 - MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
7XC3
 
 | |
