8TIX
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIR
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 5' terminal phosphates. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIU
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIS
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*A)-3'), DNA(5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-')|, MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIP
 
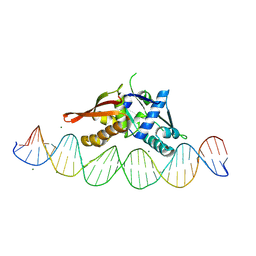 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIW
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIV
 
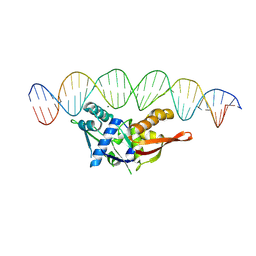 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIY
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*A)-3'), DNA(5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-')|, MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TJ1
 
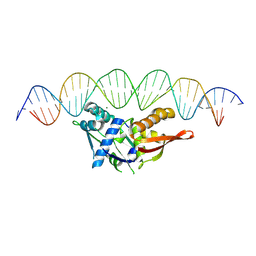 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIQ
 
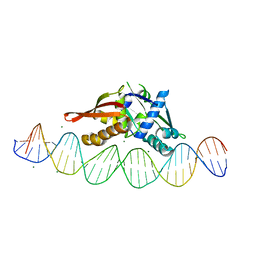 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIT
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and no terminal phosphates. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIZ
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and no terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TJ0
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
7ZGE
 
 | |
7ZF0
 
 | | Crystal structure of UGT85B1 from Sorghum bicolor in complex with UDP and p-hydroxymandelonitrile | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, 1,2-ETHANEDIOL, Cyanohydrin beta-glucosyltransferase, ... | | Authors: | Putkaradze, N, Fredslund, F, Welner, D.H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis.
Plant J., 111, 2022
|
|
7ZIE
 
 | |
7ZJV
 
 | |
7ZJU
 
 | |
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
8A1H
 
 | | Bacterial 6-4 photolyase from Vibrio cholerase | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | Authors: | Essen, L.-O, Emmerich, H.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
8A7N
 
 | | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125) | | Descriptor: | N-(3-azanylpropyl)-3-[[(3S)-1-(2-fluorophenyl)-2-oxidanylidene-pyrrolidin-3-yl]amino]-N-methyl-benzamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
