4CEK
 
 | | Crystal structure of the second MIF4G domain of human nonsense mediated decay factor UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 2 | | Authors: | Clerici, M, Deniaud, A, Boehm, V, Gehring, N.H, Schaffitzel, C, Cusack, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Analysis of the Three Mif4G Domains of Nonsense-Mediated Decay Factor Upf2.
Nucleic Acids Res., 42, 2014
|
|
4CHC
 
 | | Crystal structure of the N-terminal domain of the PA subunit of Thogoto virus polymerase (form 2) | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CHF
 
 | | Crystal structure of the putative cap-binding domain of the PB2 subunit of Thogoto virus polymerase (form 2) | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CHD
 
 | | Crystal structure of the '627' domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CQN
 
 | | Crystal structure of the E.coli LeuRS-tRNA complex with the non- cognate isoleucyl adenylate analogue | | Descriptor: | ESCHERICHIA COLI TRNA-LEU UAA ISOACCEPTOR, LEUCINE--TRNA LIGASE, MAGNESIUM ION, ... | | Authors: | Palencia, A, Cusack, S, Cvetesic, N, Haslaz, I, Gruic-Sovulj, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Physiological Target for Leurs Translational Quality Control is Norvaline
Embo J., 33, 2014
|
|
9QOF
 
 | | E.coli seryl-tRNA synthetase (Arm deletion mutant) bound to sulphamoyl seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cusack, S, Belrhali, H, Price, S, Leberman, R. | | Deposit date: | 2025-03-26 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Seryl-tRNA synthetase from Escherichia coli,implication of its N-terminal domain in aminoacylation activity and specificity.
Nucleic Acids Res, 22, 1994
|
|
1EJD
 
 | | Crystal structure of unliganded mura (type1) | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1EJC
 
 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1SER
 
 | | THE 2.9 ANGSTROMS CRYSTAL STRUCTURE OF T. THERMOPHILUS SERYL-TRNA SYNTHETASE COMPLEXED WITH TRNA SER | | Descriptor: | PROTEIN (SERYL-TRNA SYNTHETASE (E.C.6.1.1.11)), TRNASER | | Authors: | Biou, S, Cusack, V, Yaremchuk, A, Tukalo, M. | | Deposit date: | 1994-02-21 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser).
Science, 263, 1994
|
|
6XYA
 
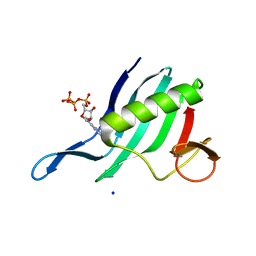 | | Cap-binding domain of SFTSV L protein | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, SODIUM ION | | Authors: | Gogrefe, N, Guenther, S, Rosenthal, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
1A4G
 
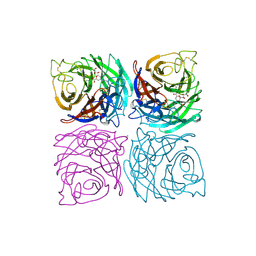 | | INFLUENZA VIRUS B/BEIJING/1/87 NEURAMINIDASE COMPLEXED WITH ZANAMIVIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | Cleasby, A, Singh, O, Skarzynski, T, Wonacott, A.J. | | Deposit date: | 1998-01-29 | | Release date: | 1999-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B.
J.Med.Chem., 41, 1998
|
|
1A4Q
 
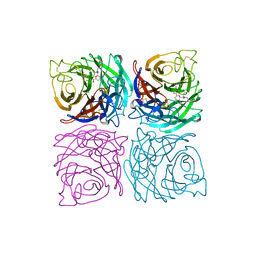 | | INFLUENZA VIRUS B/BEIJING/1/87 NEURAMINIDASE COMPLEXED WITH DIHYDROPYRAN-PHENETHYL-PROPYL-CARBOXAMIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-ACETYLAMINO-4-AMINO-6-(PHENETHYL-PROPYL-CARBAMOYL)-5,6-DIHYDRO-4H-PYRAN-2-CARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Cleasby, A, Singh, O, Skarzynski, T, Wonacott, A.J. | | Deposit date: | 1998-01-30 | | Release date: | 1999-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B.
J.Med.Chem., 41, 1998
|
|
7QUU
 
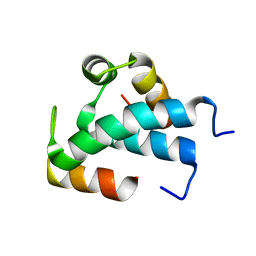 | | Red1-Iss10 complex | | Descriptor: | NURS complex subunit red1, Uncharacterized protein C7D4.14c | | Authors: | Mackereth, C.D, Kadlec, J, Laroussi, H. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
7QY5
 
 | | Crystal structure of the S.pombe Ars2-Red1 complex. | | Descriptor: | NURS complex subunit pir2, RNA elimination defective protein Red1, ZINC ION | | Authors: | Foucher, A.E, Kadlec, J. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
1V1I
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a long linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
1V1H
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a short linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
1HGJ
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGF
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGE
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
1HGD
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
2FSF
 
 | |
2FSI
 
 | |
2FSG
 
 | |
3HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
2FSH
 
 | |
