4EUO
 
 | | Structure of Atu4243-GABA sensor | | Descriptor: | ABC transporter, substrate binding protein (Polyamine), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
3OSS
 
 | | The crystal structure of enterotoxigenic Escherichia coli GspC-GspD complex from the type II secretion system | | Descriptor: | CALCIUM ION, CHLORIDE ION, TYPE 2 SECRETION SYSTEM, ... | | Authors: | Korotkov, K.V, Pruneda, J, Hol, W.G.J. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and functional studies on the interaction of GspC and GspD in the type II secretion system.
Plos Pathog., 7, 2011
|
|
3NPN
 
 | |
3PL6
 
 | |
3NPQ
 
 | |
3RCP
 
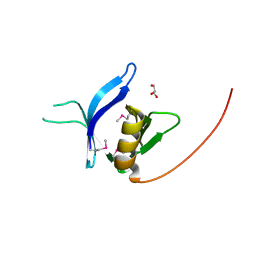 | | Crystal structure of the FAPP1 pleckstrin homology domain | | Descriptor: | GLYCEROL, Pleckstrin homology domain-containing family A member 3 | | Authors: | Roy, S, He, J, Kutateladze, T.G. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Phosphatidylinositol 4-Phosphate and ARF1 GTPase Recognition by the FAPP1 Pleckstrin Homology (PH) Domain.
J.Biol.Chem., 286, 2011
|
|
3RX8
 
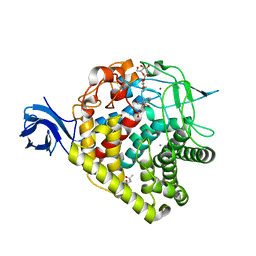 | | structure of AaCel9A in complex with cellobiose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Morera, S. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
3S1F
 
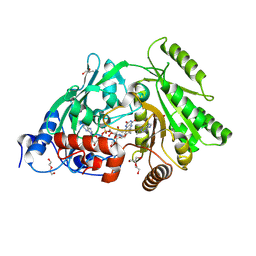 | | Asp169Glu mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3RX7
 
 | | Structure of AaCel9A in complex with cellotetraose-like isofagomine | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, CALCIUM ION, ... | | Authors: | Morera, S. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
3S1C
 
 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3K10
 
 | |
3S1D
 
 | | Glu381Ser mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3JRM
 
 | |
1Y0O
 
 | |
3MHB
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L38A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-04-07 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MVV
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS F34A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-04 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MEH
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NK9
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V74A at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-06-18 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MZ5
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L103A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MXP
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-07 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NP8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NKL
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A/I92Q at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-12 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4ODG
 
 | | Crystal structure of Staphylococcal nuclease variant V23I/V66I/V74I/V99I at cryogenic temperature | | Descriptor: | CALCIUM ION, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2014-01-10 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4NMZ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23Q/V66A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cavities in proteins
To be Published
|
|
4NDX
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92N at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Polar cavities in proteins
To be Published
|
|
