4P3Q
 
 | | Room-temperature WT DHFR, time-averaged ensemble | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Keedy, D.A, van den Bedem, H, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4P3R
 
 | | Cryogenic WT DHFR, time-averaged ensemble | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Keedy, D.A, van den Bedem, H, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
3D7K
 
 | | Crystal structure of benzaldehyde lyase in complex with the inhibitor MBP | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S. | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Probing the active center of benzaldehyde lyase with substitutions and the pseudosubstrate analogue benzoylphosphonic acid methyl ester
Biochemistry, 47, 2008
|
|
2YPI
 
 | |
3GXM
 
 | |
3GXD
 
 | | Crystal structure of Apo acid-beta-glucosidase pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GXT
 
 | |
3GXF
 
 | |
3GXP
 
 | | Crystal structure of acid-alpha-galactosidase A complexed with galactose at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GXI
 
 | | Crystal structure of acid-beta-glucosidase at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
3GXN
 
 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
1YPI
 
 | |
1O76
 
 | | CYANIDE COMPLEX OF P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYANIDE ION, ... | | Authors: | Fedorov, R, Ghosh, D, Schlichting, I. | | Deposit date: | 2002-10-23 | | Release date: | 2002-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Cyanide Complexes of P450Cam and the Oxygenase Domain of Inducible Nitric Oxide Synthase-Structural Models of the Short-Lived Oxygen Complexes
Arch.Biochem.Biophys., 409, 2003
|
|
1VSB
 
 | | SUBTILISIN CARLSBERG L-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1AVT
 
 | | SUBTILISIN CARLSBERG D-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-19 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1AV7
 
 | | SUBTILISIN CARLSBERG L-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-29 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
3VSB
 
 | | SUBTILISIN CARLSBERG D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1JPZ
 
 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
1FTX
 
 | |
4PSY
 
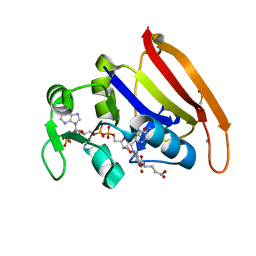 | | 100K crystal structure of Escherichia coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennet, B.C, Dealwis, C, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 22, 2014
|
|
1VGC
 
 | | GAMMA-CHYMOTRYPSIN L-PARA-CHLORO-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | GAMMA CHYMOTRYPSIN, L-1-(4-CHLOROPHENYL)-2-(ACETAMIDO)ETHANE BORONIC ACID, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
3VGC
 
 | | GAMMA-CHYMOTRYPSIN L-NAPHTHYL-1-ACETAMIDO BORONIC ACID ACID INHIBITOR COMPLEX | | Descriptor: | GAMMA CHYMOTRYPSIN, L-1-NAPHTHYL-2-ACETAMIDO-ETHANE BORONIC ACID, SULFATE ION | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1RZY
 
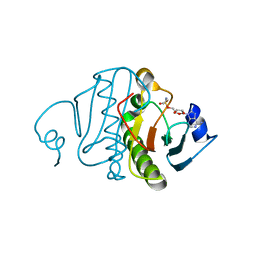 | | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | | Descriptor: | 5'-O-(N-ETHYL-SULFAMOYL)ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Krakowiak, A.K, Pace, H.C, Blackburn, G.M, Adams, M, Mekhalfia, A, Kaczmarek, R, Baraniak, J, Stec, W.J, Brenner, C. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors
J.Biol.Chem., 279, 2004
|
|
1YNO
 
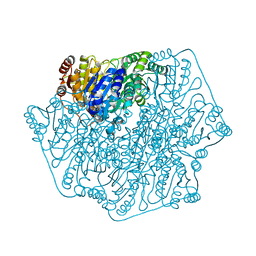 | |
1MCZ
 
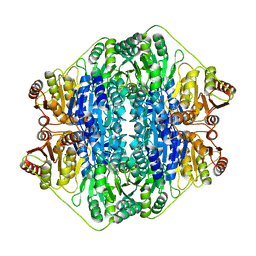 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA COMPLEXED WITH AN INHIBITOR, R-MANDELATE | | Descriptor: | (R)-MANDELIC ACID, BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Polovnikova, E.S, Bera, A.K, Hasson, M.S. | | Deposit date: | 2002-08-06 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Kinetic Analysis of Catalysis by a Thiamin
Diphosphate-Dependent Enzyme, Benzoylformate Decarboxylase
Biochemistry, 42, 2003
|
|
