8I1Y
 
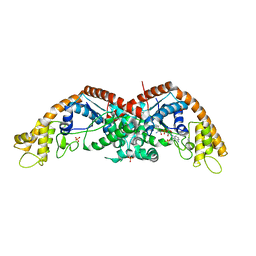 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I2A
 
 | |
8I2M
 
 | |
8I1W
 
 | |
8I2C
 
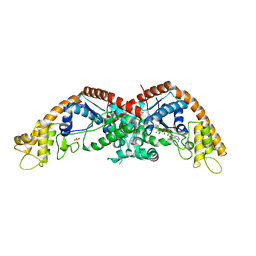 | |
8I1Z
 
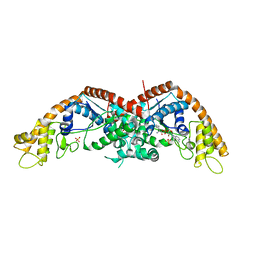 | | E. coli tryptophanyl-tRNA synthetase bound with a chemical fragment | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)ethanone, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I2L
 
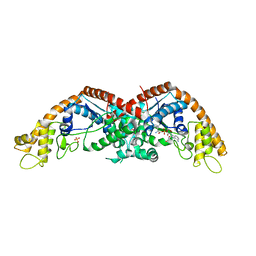 | |
7VUA
 
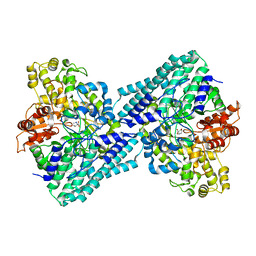 | | Anaerobic hydroxyproline degradation involving C-N cleavage by a glycyl radical enzyme | | Descriptor: | (4S)-4-hydroxy-D-proline, HplG | | Authors: | Duan, Y, Lu, Q, Yuchi, Z, Zhang, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Anaerobic Hydroxyproline Degradation Involving C-N Cleavage by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7VF6
 
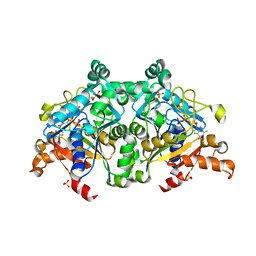 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
7F2D
 
 | |
7F2I
 
 | |
