4WJU
 
 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJS
 
 | |
3HYB
 
 | |
6YXJ
 
 | |
7O9F
 
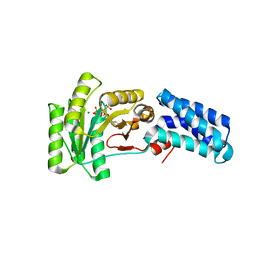 | | Bacillus subtilis Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9I
 
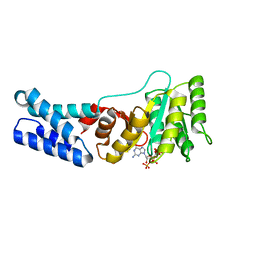 | | Escherichia coli Ffh in complex with pppGpp | | Descriptor: | Signal recognition particle protein, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9G
 
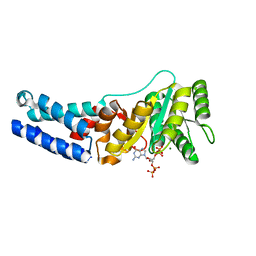 | | Escherichia coli Ffh in complex with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Signal recognition particle protein | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
7O9H
 
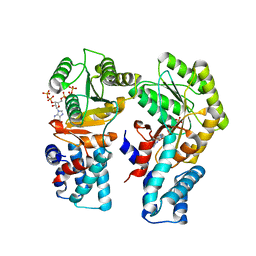 | | Escherichia coli FtsY in complex with pppGpp | | Descriptor: | Signal recognition particle receptor FtsY, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Czech, L, Mais, C.-N, Bange, G. | | Deposit date: | 2021-04-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of SRP-dependent protein secretion by the bacterial alarmone (p)ppGpp.
Nat Commun, 13, 2022
|
|
8QL1
 
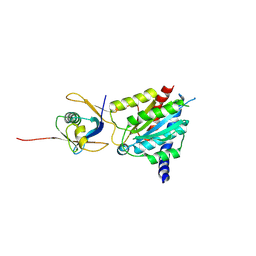 | | Crystal structure of the human MDN1-MIDAS/NLE1-UBL complex | | Descriptor: | MAGNESIUM ION, NDE1, Notchless protein homolog 1 | | Authors: | Wild, K, Fiorentino, F, Hurt, E, Sinning, I. | | Deposit date: | 2023-09-19 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly conserved ribosome biogenesis pathways between human and yeast revealed by the MDN1-NLE1 interaction and NLE1 containing pre-60S subunits.
Nucleic Acids Res., 53, 2025
|
|
3RG6
 
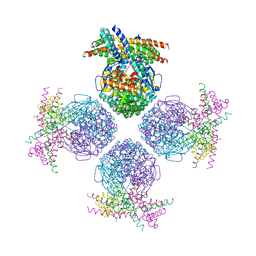 | | Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco | | Descriptor: | RbcX protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Bracher, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7AXZ
 
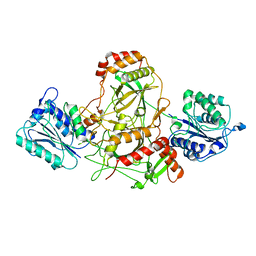 | | Ku70/80 complex apo form | | Descriptor: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | Deposit date: | 2020-11-10 | | Release date: | 2021-02-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
