1S0D
 
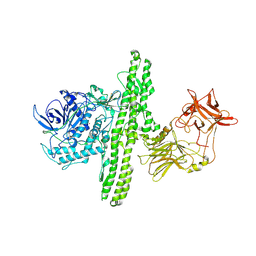 | | Crystal structure of botulinum neurotoxin type B at pH 5.5 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0G
 
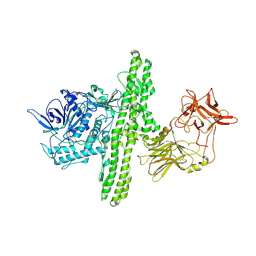 | |
1S0F
 
 | | Crystal structure of botulinum neurotoxin type B at pH 7.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1BZV
 
 | | [D-ALAB26]-DES(B27-B30)-INSULIN-B26-AMIDE A SUPERPOTENT SINGLE-REPLACEMENT INSULIN ANALOGUE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INSULIN | | Authors: | Kurapkat, G, Siedentopf, M, Gattner, H.G, Hagelstein, M, Brandenburg, D, Grotzinger, J, Wollmer, A. | | Deposit date: | 1998-11-04 | | Release date: | 1999-05-18 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a superpotent B-chain-shortened single-replacement insulin analogue.
Protein Sci., 8, 1999
|
|
1S0E
 
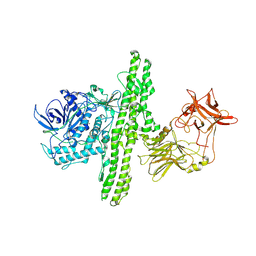 | | Crystal structure of botulinum neurotoxin type B at pH 6.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
2HFV
 
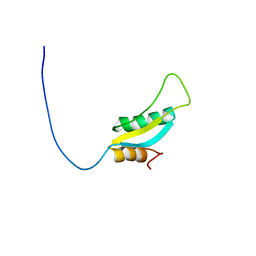 | | Solution NMR Structure of Protein RPA1041 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT90. | | Descriptor: | Hypothetical Protein RPA1041 | | Authors: | Eletsky, A, Atreya, H.S, Liu, G, Sukumaran, D, Garcia, M, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Pseudomonas aeruginosa Hypothetical Protein RPA1041
TO BE PUBLISHED
|
|
2KAN
 
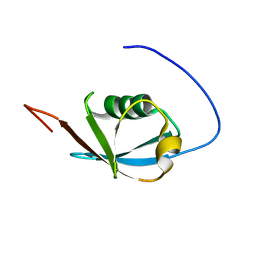 | | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein ar3433a | | Authors: | Eletsky, A, Mills, J.L, Sukumaran, D, Hua, J, Shastry, R, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Everret, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-11 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350.
To be Published
|
|
2K5H
 
 | | Solution NMR structure of protein encoded by MTH693 from Methanobacterium thermoautotrophicum: Northeast Structural Genomics Consortium target tt824a | | Descriptor: | Conserved protein | | Authors: | Wu, Y, Singarapu, K, Semesi, A, Sukumaran, D, Yee, A, Garcia, M, Arrowsmith, C, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein encoded by MTH693 from Methanobacterium thermoautotrophicum: Northeast Structural Genomics Consortium target tt824a
To be Published
|
|
2K4M
 
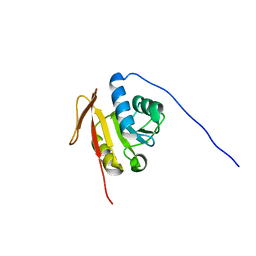 | | Solution NMR Structure of M. thermoautotrophicum protein MTH_1000, Northeast Structural Genomics Consortium Target TR8 | | Descriptor: | UPF0146 protein MTH_1000 | | Authors: | Eletsky, A, Sukumaran, D, Semesi, A, Guido, V, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of M. thermoautotrophicum protein MTH_1000, Northeast Structural Genomics Consortium Target TR8
To be Published
|
|
5BUW
 
 | |
2KEL
 
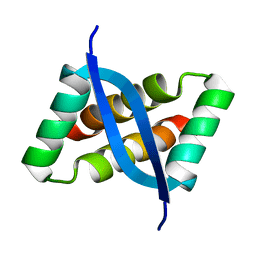 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
1TH4
 
 | | crystal structure of NADPH depleted bovine liver catalase complexed with 3-amino-1,2,4-triazole | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugadev, R, Ponnuswamy, M.N, Kumaran, D, Swaminathan, S, Sekar, K. | | Deposit date: | 2004-06-01 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | crystal structure of bovine liver catalase
TO BE PUBLISHED
|
|
3BCV
 
 | |
2GOK
 
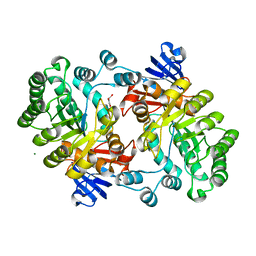 | | Crystal structure of the imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution | | Descriptor: | CHLORIDE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Tyagi, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-13 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
2KKS
 
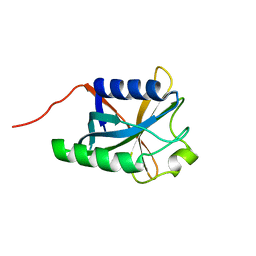 | | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, Y, Mills, J.L, Wang, H, Ciccosanti, C, Jiang, M, Sukumaran, D, Zhang, Q, Nair, R, Rost, B, Acton, T, Xiao, R, Swapna, G.V.T, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27
To be Published
|
|
2KC7
 
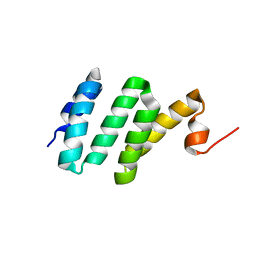 | | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218 | | Descriptor: | bfr218_protein | | Authors: | Eletsky, A, Wu, Y, Sukumaran, D, Lee, H, Lee, D.Y, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-17 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218
To be Published
|
|
5BUY
 
 | |
1PV1
 
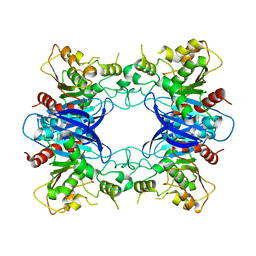 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | Descriptor: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | Authors: | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-26 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
2GU1
 
 | | Crystal structure of a zinc containing peptidase from vibrio cholerae | | Descriptor: | SODIUM ION, ZINC ION, Zinc peptidase | | Authors: | Sugadev, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae.
Proteins, 72, 2008
|
|
2KCQ
 
 | | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855) . Northeast Structural Genomics Consortium target SrR106 | | Descriptor: | Mov34/MPN/PAD-1 family | | Authors: | Wu, Y, Eletsky, A, Zhao, L, Hua, J, Sukumaran, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-28 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855). Northeast Structural Genomics Consortium target SrR106
To be Published
|
|
2HAF
 
 | | Crystal structure of a putative translation repressor from Vibrio cholerae | | Descriptor: | Putative translation repressor | | Authors: | Sugadev, R, Seetharaman, J, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of a putative translation repressor from Vibrio cholerae
To be Published
|
|
1QJZ
 
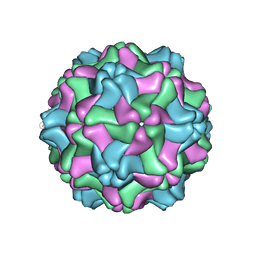 | | Three Dimensional Structure of Physalis Mottle Virus : Implications for the Viral Assembly | | Descriptor: | COAT PROTEIN | | Authors: | Krishna, S.S, Hiremath, C.N, Munshi, S.K, Prahadeeswaran, D, Sastri, M, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three Dimensional Structure of Physalis Movirus: Implications for the Viral Assembly
J.Mol.Biol., 289, 1999
|
|
5BUX
 
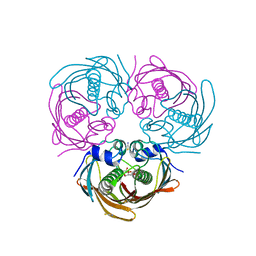 | |
6NAV
 
 | | Cryo-EM reconstruction of Sulfolobus islandicus LAL14/1 Pilus | | Descriptor: | M9UD72 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2018-12-06 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | An extensively glycosylated archaeal pilus survives extreme conditions.
Nat Microbiol, 4, 2019
|
|
3BZW
 
 | | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron | | Descriptor: | ACETATE ION, Putative lipase, SULFATE ION | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron.
To be Published
|
|
