8BTE
 
 | | Notum Inhibitor ARUK3004470 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSQ
 
 | | Notum Inhibitor ARUK3006561 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT0
 
 | | Notum Inhibitor ARUK3005518 | | Descriptor: | (3~{a}~{S})-2,2-bis(fluoranyl)-3~{a},4-dihydro-3~{H}-pyrrolo[1,2-a]indol-1-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSZ
 
 | | Notum Inhibitor ARUK3005522 | | Descriptor: | (3~{a}~{R})-2,3,3~{a},4-tetrahydropyrrolo[1,2-a]indol-1-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTA
 
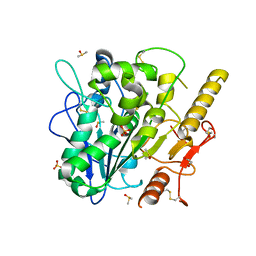 | | Notum Inhibitor ARUK3004308 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chloranyl-2,3-dihydroindol-1-yl)ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTH
 
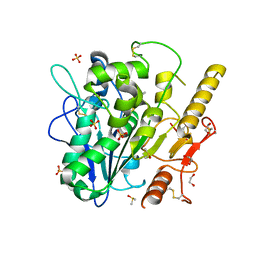 | | Notum Inhibitor ARUK3004552 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydroindol-1-yl)prop-2-en-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT8
 
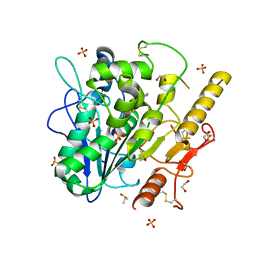 | | Notum Inhibitor ARUK3004048 | | Descriptor: | 1,2-ETHANEDIOL, 1-indol-1-ylethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-28 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
5YJ3
 
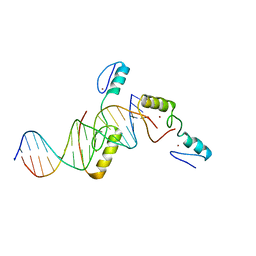 | | Crystal structure of TZAP and telomeric DNA complex | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), Telomere zinc finger-associated protein, ... | | Authors: | Li, F, Zhao, Y. | | Deposit date: | 2017-10-07 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA.
Cell Res., 28, 2018
|
|
4RDP
 
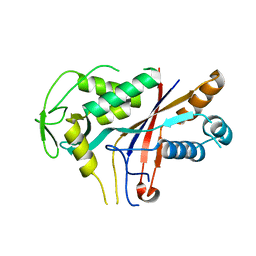 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
5E08
 
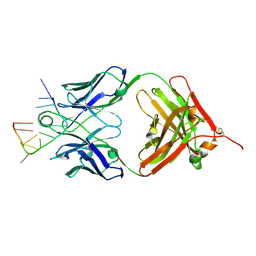 | | Specific Recognition of a Single-stranded RNA Sequence by an Engineered Synthetic Antibody Fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, RNA | | Authors: | Huang, H, Qin, D, Li, N, Shao, Y, Staley, J.P, Kossiakoff, A.A, Koide, S, Piccirilli, J.A. | | Deposit date: | 2015-09-28 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment.
J.Mol.Biol., 428, 2016
|
|
5VOV
 
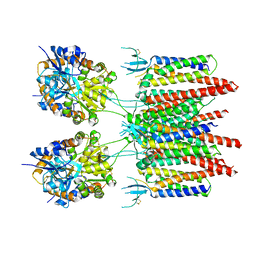 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
6NJL
 
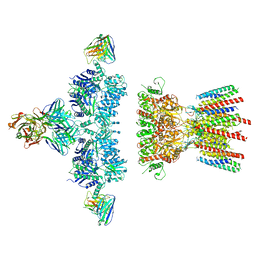 | |
6NJM
 
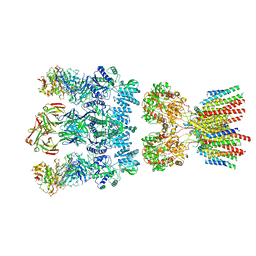 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
6NJN
 
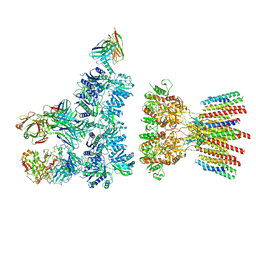 | |
7TUB
 
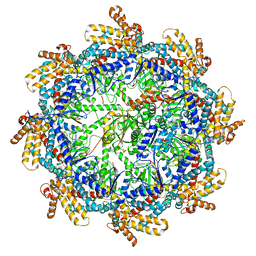 | | The beta-tubulin folding intermediate IV | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-02 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TRG
 
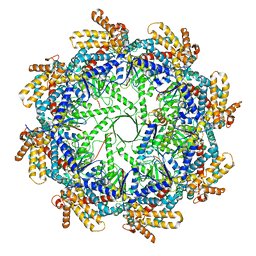 | | The beta-tubulin folding intermediate I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTN
 
 | | The beta-tubulin folding intermediate II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTT
 
 | | The beta-tubulin folding intermediate III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
8JGT
 
 | |
8JGR
 
 | |
8JGW
 
 | |
8JGX
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
