8SMT
 
 | | Crystal structure of antibody WRAIR-2134 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2134 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
4XXC
 
 | | HLA-B*1801 in complex with a self-peptide, DELEIKAY | | Descriptor: | ACETATE ION, ASP-GLU-LEU-GLU-ILE-LYS-ALA-TYR, Beta-2-microglobulin, ... | | Authors: | Hibbert, K.M, Rossjohn, J, Gras, S. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.426 Å) | | Cite: | T Cell Cross-Reactivity between a Highly Immunogenic EBV Epitope and a Self-Peptide Naturally Presented by HLA-B*18:01+ Cells.
J Immunol., 194, 2015
|
|
4YZU
 
 | |
8SMI
 
 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SGU
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-13 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
4Z0K
 
 | |
1FGS
 
 | | FOLYLPOLYGLUTAMATE SYNTHETASE FROM LACTOBACILLUS CASEI | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHETASE, MAGNESIUM ION, PYROPHOSPHATE 2- | | Authors: | Sun, X, Bognar, A, Baker, E, Smith, C. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7TLT
 
 | | SARS-CoV-2 Spike-derived peptide S489-497 (YFPLQSYGF) presented by HLA-A*29:02 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Murdolo, L.D, Szeto, C, Gras, S. | | Deposit date: | 2022-01-18 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ablation of CD8 + T cell recognition of an immunodominant epitope in SARS-CoV-2 Omicron variants BA.1, BA.2 and BA.3.
Nat Commun, 13, 2022
|
|
7RTD
 
 | |
7RTR
 
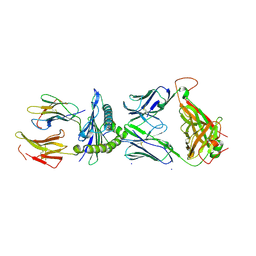 | |
7KGS
 
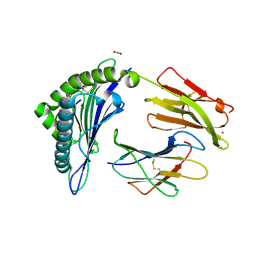 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N138-146 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGT
 
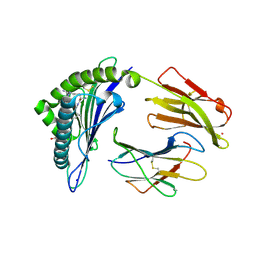 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N226-234 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGR
 
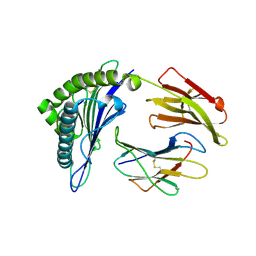 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N159-167 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nucleoprotein | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGQ
 
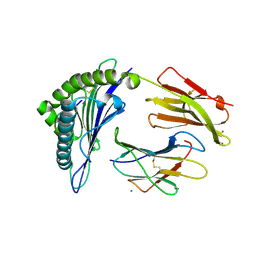 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N222-230 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGP
 
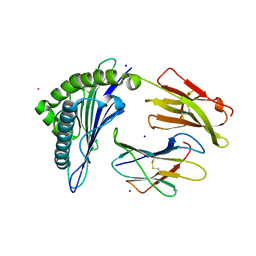 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGO
 
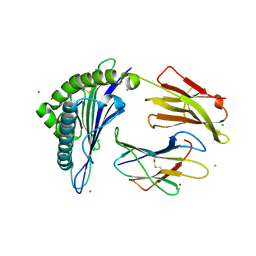 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
2WLK
 
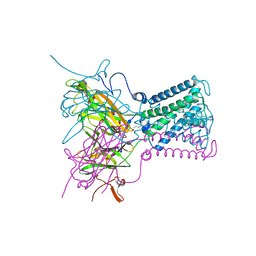 | | STRUCTURE OF THE ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL FROM MAGNETOSPIRILLUM MAGNETOTACTICUM | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2WLJ
 
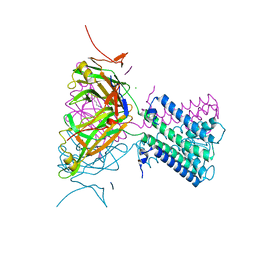 | | Potassium channel from Magnetospirillum magnetotacticum | | Descriptor: | CALCIUM ION, CHLORIDE ION, POTASSIUM CHANNEL, ... | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
6C0W
 
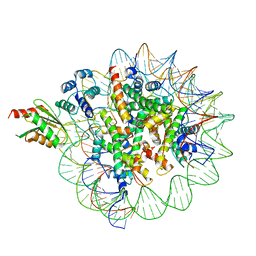 | | Cryo-EM structure of human kinetochore protein CENP-N with the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Centromere protein N, Histone H2A, ... | | Authors: | Zhou, K, Pentakota, S, Vetter, I.R, Morgan, G.P, Petrovic, A, Musacchio, A, Luger, K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Decoding the centromeric nucleosome through CENP-N.
Elife, 6, 2017
|
|
1FC5
 
 | |
6EQT
 
 | |
4ZVJ
 
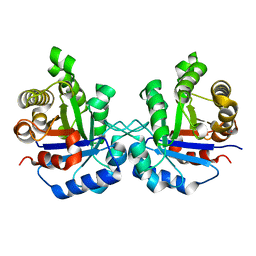 | | Structure of human triose phosphate isomerase K13M | | Descriptor: | POTASSIUM ION, SODIUM ION, Triosephosphate isomerase | | Authors: | Amrich, C.G, Smith, C, Heroux, A, VanDemark, A.P. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6996 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim. Biophys. Acta, 1852, 2015
|
|
3LEZ
 
 | |
3VCL
 
 | |
3LZ6
 
 | | Guinea Pig 11beta hydroxysteroid dehydrogenase with PF-877423 | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-adamantan-2-yl-1-ethyl-D-prolinamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pauly, T.A. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The development and SAR of pyrrolidine carboxamide 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
