6ZHS
 
 | | Uba1 bound to two E2 (Ubc13) molecules | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHT
 
 | | Uba1-Ubc13 disulfide mediated complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Schaefer, A, Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHU
 
 | | Yeast Uba1 in complex with Ubc3 and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system
To Be Published
|
|
7VL4
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with methyl beta-D-glucoside | | Descriptor: | CALCIUM ION, beta-1,2-glucosyltransferase, methyl beta-D-glucopyranoside | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
7VL1
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with methyl alpha-D-glucoside | | Descriptor: | CALCIUM ION, beta-1,2-glucosyltransferase, methyl alpha-D-glucopyranoside | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
1A05
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
7X87
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophotetraose observed as sophorose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J Biol Chem, 298, 2022
|
|
1WR1
 
 | | The complex structure of Dsk2p UBA with ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin-like protein DSK2 | | Authors: | Ohno, A, Jee, J.G, Fujiwara, K, Tenno, T, Goda, N, Tochio, H, Hiroaki, H, kobayashi, H, Shirakawa, M. | | Deposit date: | 2004-10-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-09-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition.
Structure, 13, 2005
|
|
7PYV
 
 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7PVN
 
 | | Crystal Structure of Human UBA6 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Truongvan, N, Li, S, Schindelin, H. | | Deposit date: | 2021-10-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
1WR0
 
 | | Structural characterization of the MIT domain from human Vps4b | | Descriptor: | SKD1 protein | | Authors: | Takasu, H, Jee, J.G, Ohno, A, Goda, N, Fujiwara, K, Tochio, H, Shirakawa, M, Hiroaki, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-07 | | Release date: | 2005-08-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the MIT domain from human Vps4b
Biochem.Biophys.Res.Commun., 334, 2005
|
|
7PO7
 
 | | Phosphoglycolate phosphatase from Mus musculus | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Schloetzer, J, Schindelin, H, Fratz, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycolytic flux control by drugging phosphoglycolate phosphatase.
Nat Commun, 13, 2022
|
|
7PUX
 
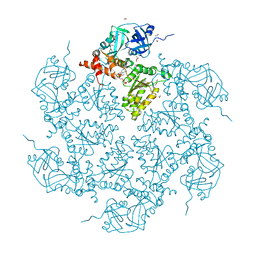 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
7POE
 
 | | Phosphoglycolate Phosphatase with Inhibitor CP1 | | Descriptor: | 2-[[4-[4-[(2-carboxyphenyl)carbamoyl]phenoxy]phenyl]carbonylamino]benzoic acid, GLYCEROL, Glycerol-3-phosphate phosphatase, ... | | Authors: | Schloetzer, J, Fratz, S, Schindelin, H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Glycolytic flux control by drugging phosphoglycolate phosphatase.
Nat Commun, 13, 2022
|
|
7PVP
 
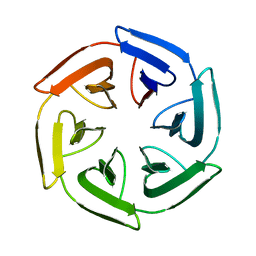 | | Crystal structure of the SAKe6BC designer protein | | Descriptor: | SAKe6BC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-10-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7Q58
 
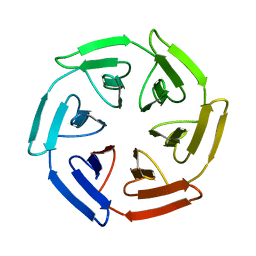 | | Crystal structure of the SAKe6BR designer protein | | Descriptor: | SAKe6BR | | Authors: | Wouters, S.M.L, Noguchi, H, Velupla, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-11-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
1NVJ
 
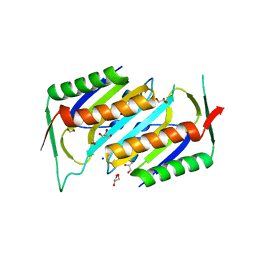 | | Deletion Mutant (Delta 141) of Molybdopterin Synthase | | Descriptor: | FORMIC ACID, GLYCEROL, Molybdopterin converting factor subunit 2, ... | | Authors: | Rudolph, M.J, Wuebbens, M.M, Turque, O, Rajagopalan, K.V, Schindelin, H. | | Deposit date: | 2003-02-03 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Studies of Molybdopterin Synthase Provide Insights into its Catalytic Mechanism
J.Biol.Chem., 278, 2003
|
|
7OV7
 
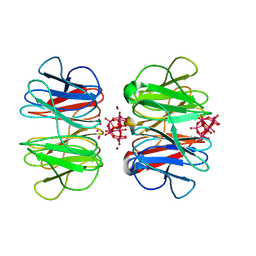 | | The hybrid cage formed between Pizza6-S and Cu(II)-substituted trilacunary Keggin | | Descriptor: | COPPER (II) ION, POTASSIUM ION, Pizza6-S, ... | | Authors: | Vandebroek, L, Noguchi, H, Anyushin, A, Van Meervelt, L, Voet, A.R.D, Parac-Vogt, T.N. | | Deposit date: | 2021-06-14 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hierarchical Self-Assembly of a Supramolecular Protein-Metal Cage Encapsulating a Polyoxometalate Guest
Cryst.Growth Des., 2022
|
|
7ONG
 
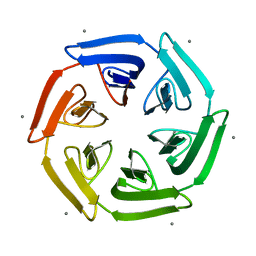 | | Crystal structure of the computationally designed SAKe6BE-L1 protein | | Descriptor: | CALCIUM ION, SAKe6BE-L1 | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7OPU
 
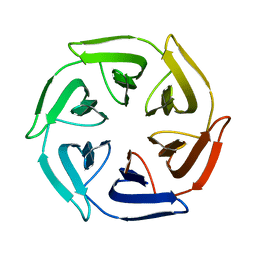 | |
7ON7
 
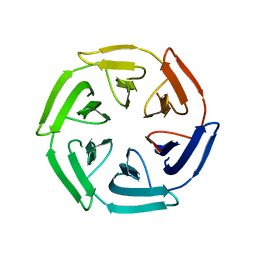 | |
7ON8
 
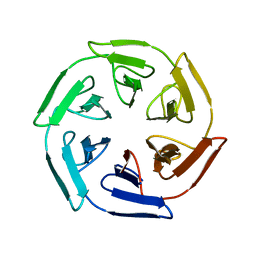 | |
7ONA
 
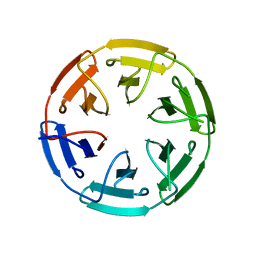 | | Crystal structure of the computationally designed SAKe6AC protein | | Descriptor: | CALCIUM ION, SAKe6AC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONE
 
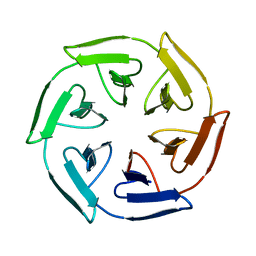 | | Crystal structure of the self-assembled SAKe6BE designer protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONC
 
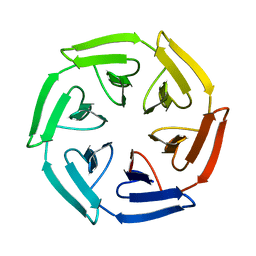 | | Crystal structure of the computationally designed SAKe6BE protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
