7T7U
 
 | | Light Harvesting complex phycocyanin PC 630, from the cryptophyte Chroomonas sp. M1627 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOCYANOBILIN, ... | | Authors: | Michie, K.A, Harrop, S.J, Rathbone, H.W, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2021-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
7T89
 
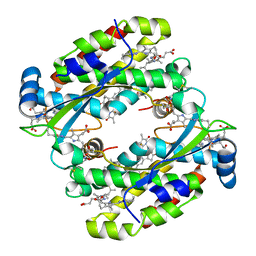 | | Light harvesting complex Phycocyanin PC577 from the cryptophyte Hemiselmis pacifica CCMP 706 | | Descriptor: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Michie, K.A, Curmi, P.C, Harrop, S, Rathbone, H.W. | | Deposit date: | 2021-12-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Molecular structures reveal the origin of spectral variation in cryptophyte light harvesting antenna proteins.
Protein Sci., 32, 2023
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7TN1
 
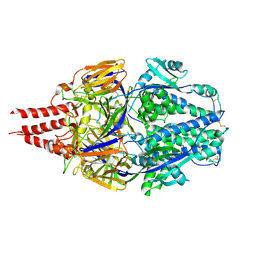 | | Multistate design to stabilize viral class I fusion proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Huang, J, Banerjee, A, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-01-20 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Nat Commun, 15, 2024
|
|
8AKL
 
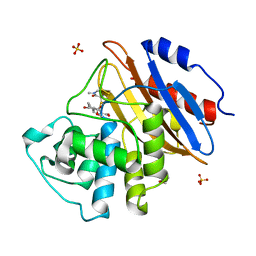 | | Acyl-enzyme complex of meropenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2S,3R,4R)-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl-3-methyl-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
7U7O
 
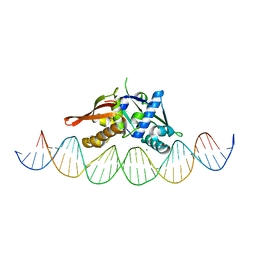 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional T-A rich sequence. | | Descriptor: | DNA (5'-D(A*TP*AP*AP*TP*TP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*TP*AP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*AP*AP*T)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2022-03-07 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UFX
 
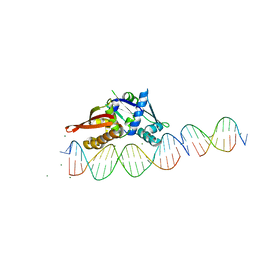 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold-insert duplexes (21mer and 10mer) containing the cognate REPE54 sequence and an additional G-C rich sequence, respectively. | | Descriptor: | DNA (5'-D(A*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(A*GP*AP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(A*GP*TP*CP*CP*GP*GP*GP*CP*CP*G)-3'), ... | | Authors: | Ward, A.R, Shields, E.T, Snow, C.D. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7U6K
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UOG
 
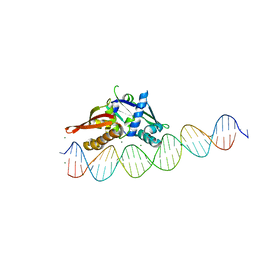 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and asymmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*CP*CP*GP*CP*CP*TP*T)-3', DNA (5'-D(GP*TP*AP*AP*GP*GP*CP*GP*GP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UR0
 
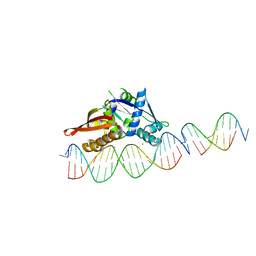 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold-insert duplexes (21mer and 10mer) containing the cognate REPE54 sequence and an additional T-A rich sequence, respectively. | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(A*GP*AP*CP*GP*GP*TP*AP*AP*TP*T)-3'), DNA (5'-D(A*GP*TP*AP*AP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UXY
 
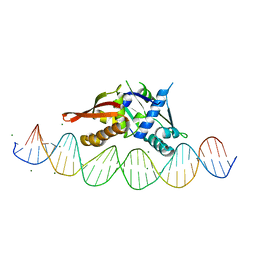 | | Isoreticular, interpenetrating co-crystal of protein variant Replication Initiator Protein REPE54 (L53G,Q54G,E55G) and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*P*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*P*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8DSS
 
 | |
7TUR
 
 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
8DTA
 
 | |
8E2N
 
 | |
8AKK
 
 | | Acyl-enzyme complex of imipenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKI
 
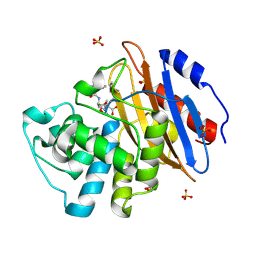 | | Acyl-enzyme complex of ampicillin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8EUO
 
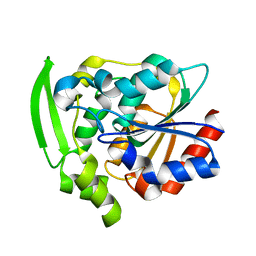 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
8AKM
 
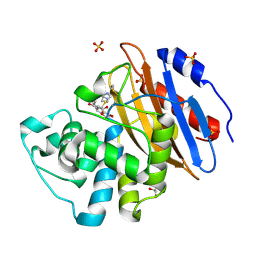 | | Acyl-enzyme complex of ertapenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, Ertapenem, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKJ
 
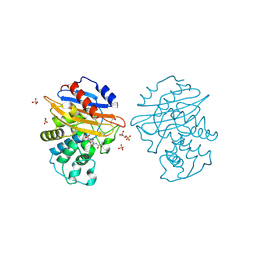 | | Acyl-enzyme complex of cephalothin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8ERX
 
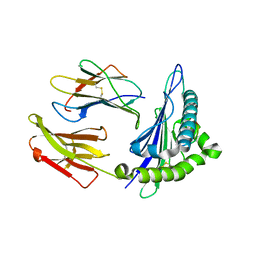 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
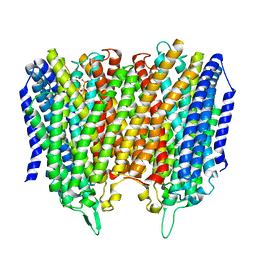
8B22
 
 | |
8B23
 
 | |
8B21
 
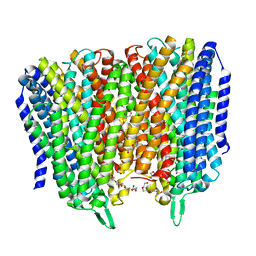 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
