3WGX
 
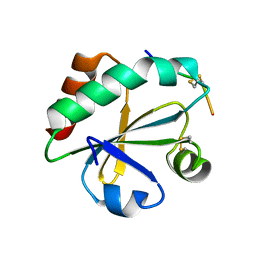 | | Crystal structure of ERp46 Trx2 in a complex with Prx4 C-term | | Descriptor: | GLYCEROL, Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-13 | | Release date: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
5YOK
 
 | | Structure of HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, HIV-1 PROTEASE | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
2Z5V
 
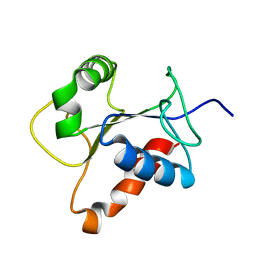 | | Solution structure of the TIR domain of human MyD88 | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Ohnishi, H, Tochio, H, Hiroaki, H, Kondo, N, Kato, Z, Shirakawa, M. | | Deposit date: | 2007-07-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling.
Proc.Natl.Acad.Sci.USA, 2009
|
|
2YR4
 
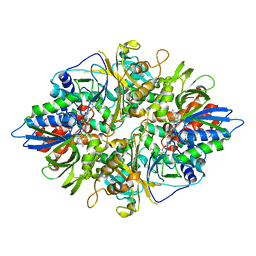 | | Crystal structure of L-phenylalanine oxiase from Psuedomonas sp. P-501 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pro-enzyme of L-phenylalanine oxidase, SULFATE ION | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2YR6
 
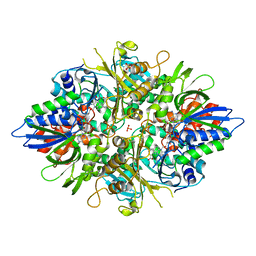 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2FCR
 
 | |
