1KMA
 
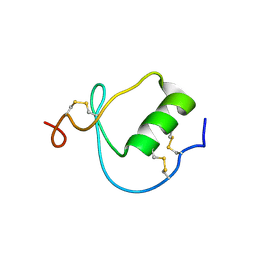 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
6GZK
 
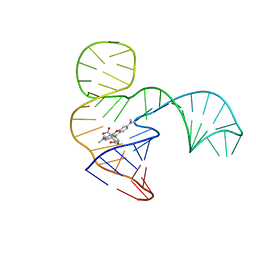 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
6GZR
 
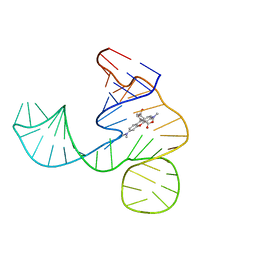 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
1RFR
 
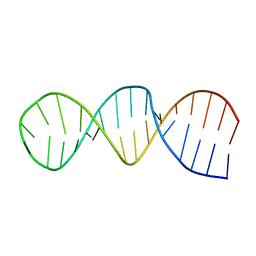 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
3BBD
 
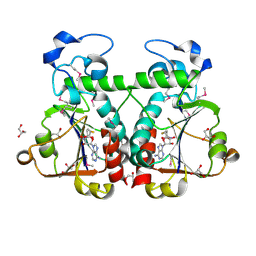 | | M. jannaschii Nep1 complexed with S-adenosyl-homocysteine | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBE
 
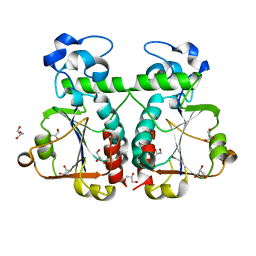 | | M. jannaschii Nep1 | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBH
 
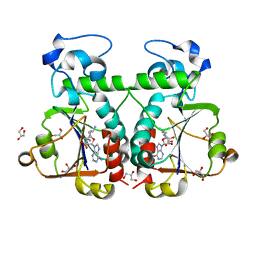 | | M. jannaschii Nep1 complexed with Sinefungin | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
1D6K
 
 | | NMR SOLUTION STRUCTURE OF THE 5S RRNA E-LOOP/L25 COMPLEX | | Descriptor: | 5S RRNA E-LOOP (5SE), RIBOSOMAL PROTEIN L25 | | Authors: | Stoldt, M, Wohnert, J, Ohlenschlager, O, Gorlach, M, Brown, L.R. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 5S rRNA E-domain-protein L25 complex shows preformed and induced recognition.
EMBO J., 18, 1999
|
|
2MXS
 
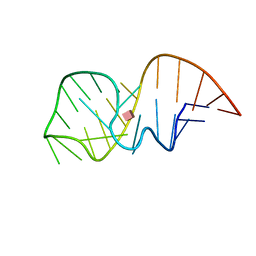 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2KXM
 
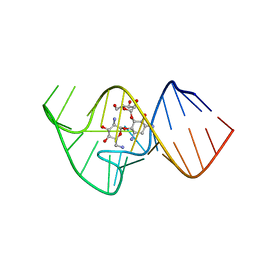 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6EWS
 
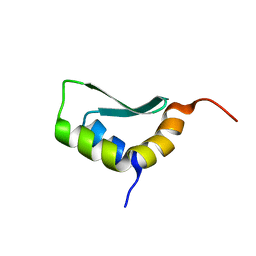 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
5AP8
 
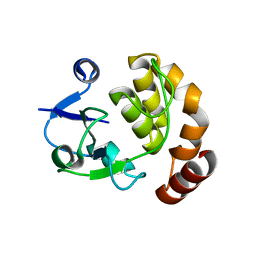 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | Descriptor: | TSR3 | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
8CQ1
 
 | |
6TRP
 
 | | Solution Structure of Docking Domain Complex of Pax NRPS: PaxC NDD - PaxB CDD | | Descriptor: | Peptide synthetase XpsB,Peptide synthetase XpsB | | Authors: | Watzel, J, Hacker, C, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A New Docking Domain Type in the Peptide-Antimicrobial-Xenorhabdus Peptide Producing Nonribosomal Peptide Synthetase fromXenorhabdus bovienii.
Acs Chem.Biol., 15, 2020
|
|
487D
 
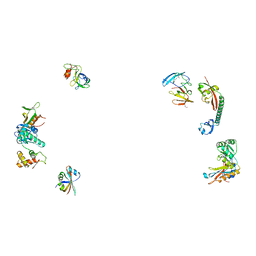 | |
3RKF
 
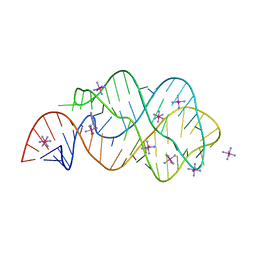 | | Crystal structure of guanine riboswitch C61U/G37A double mutant bound to thio-guanine | | Descriptor: | 2-amino-1,9-dihydro-6H-purine-6-thione, COBALT HEXAMMINE(III), Guanine riboswitch | | Authors: | Buck, J, Wacker, A, Warkentin, E, Woehnert, J, Wirmer-Bartoschek, J, Schwalbe, H. | | Deposit date: | 2011-04-18 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Influence of ground-state structure and Mg2+ binding on folding kinetics of the guanine-sensing riboswitch aptamer domain.
Nucleic Acids Res., 39, 2011
|
|
2N0J
 
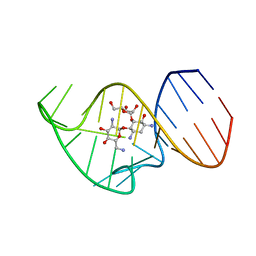 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | Descriptor: | RIBOSTAMYCIN, RNA_(27-MER) | | Authors: | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | Deposit date: | 2015-03-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8BWT
 
 | |
6S10
 
 | |
5LWJ
 
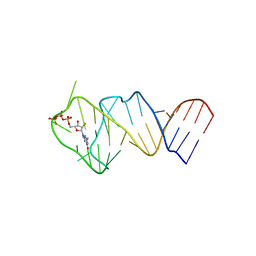 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7B2B
 
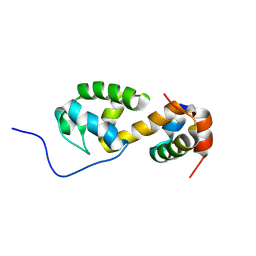 | | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | | Descriptor: | Amino acid adenylation domain-containing protein, Peptide synthetase PaxA | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B2F
 
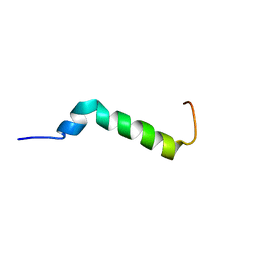 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4QVK
 
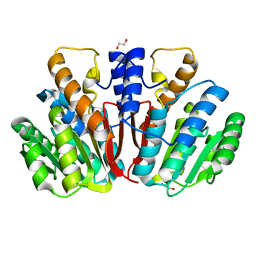 | | Apo-crystal structure of Podospora anserina methyltransferase PaMTH1 | | Descriptor: | 1,2-ETHANEDIOL, PaMTH1 Methyltransferase | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2014-07-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
6HAG
 
 | |
6EWU
 
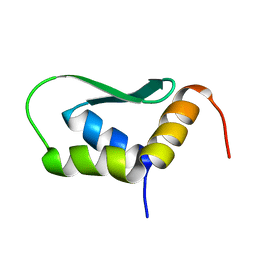 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
