3CP5
 
 | | Cytochrome c from rhodothermus marinus | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Stelter, M, Melo, A, Saraiva, L, Teixeira, M, Archer, M. | | Deposit date: | 2008-03-31 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A novel type of monoheme cytochrome c: biochemical and structural characterization at 1.23 A resolution of rhodothermus marinus cytochrome c
Biochemistry, 47, 2008
|
|
3H31
 
 | | Structure of Rhodothermus marinus HiPIP at 1.0 A resolution | | Descriptor: | High potential iron-sulfur protein, IRON/SULFUR CLUSTER | | Authors: | Stelter, M, Melo, A.M.P, Saraiva, L, Teixeira, M, Archer, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-12-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure at 1.0 A resolution of a high-potential iron-sulfur protein involved in the aerobic respiratory chain of Rhodothermus marinus
J.Biol.Inorg.Chem., 15, 2010
|
|
4A1F
 
 | | Crystal structure of C-terminal domain of Helicobacter pylori DnaB Helicase | | Descriptor: | CITRATE ANION, REPLICATIVE DNA HELICASE | | Authors: | Stelter, M, Kapp, U, Timmins, J, Terradot, L. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of a Dodecameric Bacterial Replicative Helicase.
Structure, 20, 2012
|
|
4C2T
 
 | | Crystal structure of full length Deinococcus radiodurans UvrD in complex with DNA | | Descriptor: | DNA HELICASE II, DNA STRAND FOR28, DNA STRAND REV28, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.997 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4C2U
 
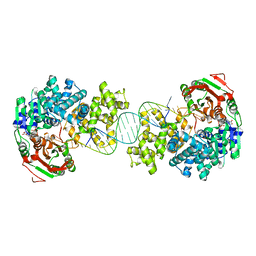 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, Form 1 | | Descriptor: | DNA HELICASE II, FOR25, GLYCEROL, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4C30
 
 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, form 2 | | Descriptor: | DNA HELICASE II, DNA STRAND FOR25, DNA STRAND REV25, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4LXR
 
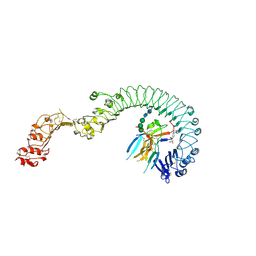 | | Structure of the Toll - Spatzle complex, a molecular hub in Drosophila development and innate immunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stelter, M, Parthier, C, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2013-07-30 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4LXS
 
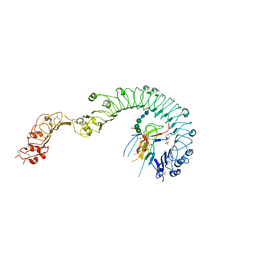 | | Structure of the Toll - Spatzle complex, a molecular hub in Drosophila development and innate immunity (glycosylated form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stelter, M, Parthier, C, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2013-07-30 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Toll-Spatzle complex, a molecular hub in Drosophila development and innate immunity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8B0Q
 
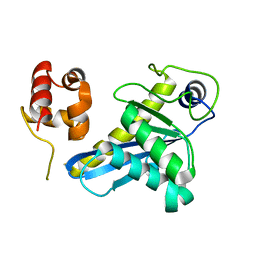 | | Deinococcus radiodurans UvrC C-terminal half | | Descriptor: | UvrABC system protein C | | Authors: | Timmins, J, Stelter, M. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.
Nucleic Acids Res., 51, 2023
|
|
3H8L
 
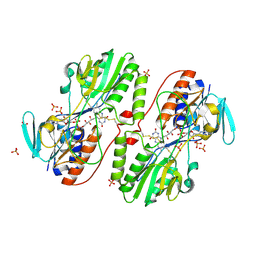 | | The first X-ray structure of a sulfide:quinone oxidoreductase: insights into sulfide oxidation mechanism | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase, PHOSPHATE ION, ... | | Authors: | Brito, J.A, Sousa, F.L, Stelter, M, Bandeiras, T.M, Vonrhein, C, Teixeira, M, Pereira, M.M, Archer, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into sulfide:quinone oxidoreductase.
Biochemistry, 48, 2009
|
|
3H8I
 
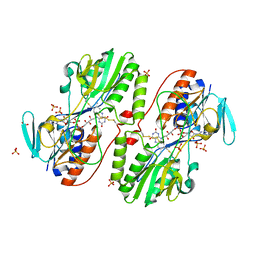 | | The first X-ray structure of a sulfide:quinone oxidoreductase: Insights into sulfide oxidation mechanism | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase, PHOSPHATE ION, ... | | Authors: | Brito, J.A, Sousa, F.L, Stelter, M, Bandeiras, T.M, Vonrhein, C, Teixeira, M, Pereira, M.M, Archer, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional insights into sulfide:quinone oxidoreductase.
Biochemistry, 48, 2009
|
|
2V05
 
 | |
2V04
 
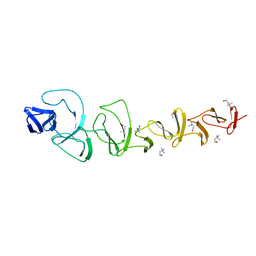 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Hermoso, J, Molina, R, Kahn, R, Stelter, M. | | Deposit date: | 2007-05-08 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
2W4E
 
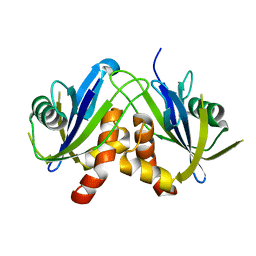 | | Structure of an N-terminally truncated Nudix hydrolase DR2204 from Deinococcus radiodurans | | Descriptor: | MUTT/NUDIX FAMILY PROTEIN | | Authors: | Goncalves, A.M.D, Fioravanti, E, Stelter, M, McSweeney, S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an N-Terminally Truncated Nudix Hydrolase Dr2204 from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1OKJ
 
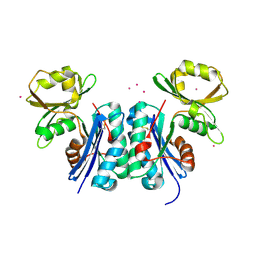 | |
2VYU
 
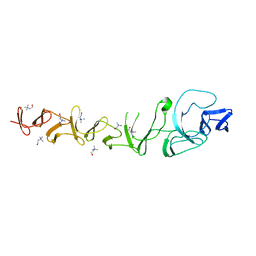 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE IN THE PRESENCE OF A PEPTIDOGLYCAN ANALOGUE (TETRASACCHARIDE-PENTAPEPTIDE) | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Perez-Dorado, I, Molina, R, Hermoso, J.A, Mobashery, S. | | Deposit date: | 2008-07-28 | | Release date: | 2009-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
2BIB
 
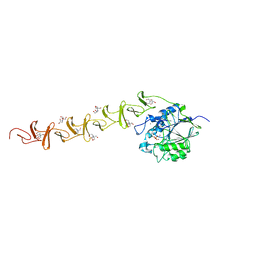 | | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, PHOSPHOCHOLINE, ... | | Authors: | Hermoso, J.A, Lagartera, L, Gonzalez, A, Garcia, P, Martinez-Ripoll, M, Garcia, J.L, Menendez, M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights Into Pneumococcal Pathogenesis from Crystal Structure of the Modular Teichoic Acid Phosphorylcholine Esterase Pce
Nat.Struct.Mol.Biol., 12, 2005
|
|
3NOM
 
 | | Crystal Structure of Zymomonas mobilis Glutaminyl Cyclase (monoclinic form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3NOL
 
 | | Crystal structure of Zymomonas mobilis Glutaminyl Cyclase (trigonal form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3NOK
 
 | | Crystal structure of Myxococcus xanthus Glutaminyl Cyclase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
